|
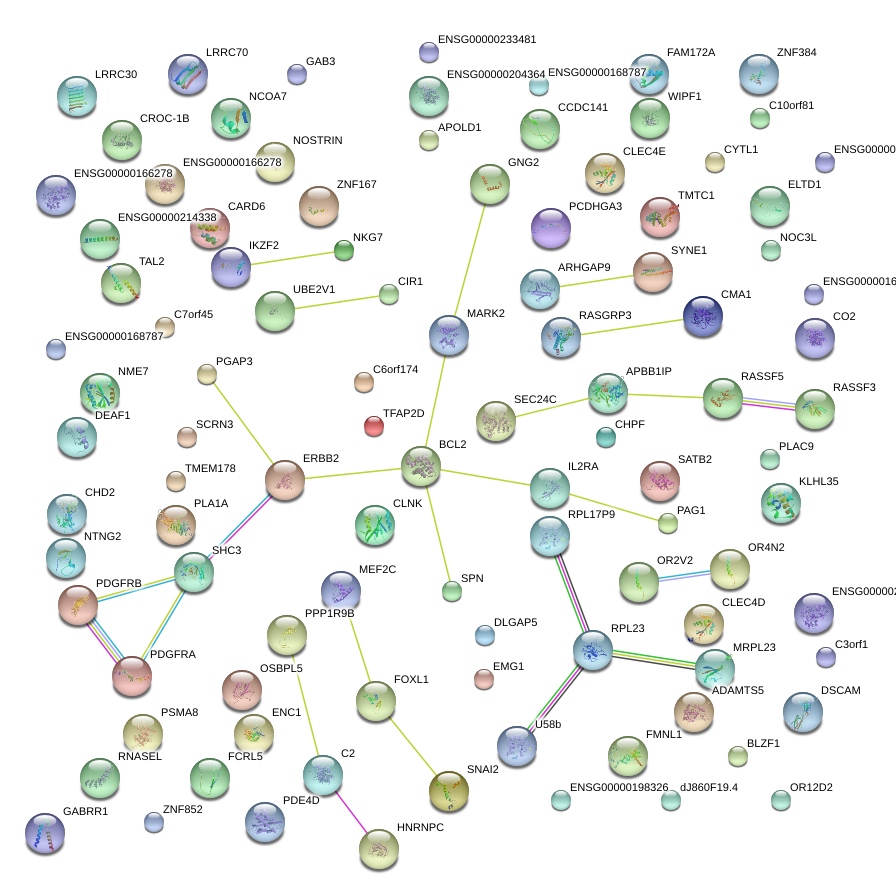
chr4_+_54790020
|
0.545
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr3_+_120799411
|
0.499
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr19_-_56567602
|
0.461
|
NM_005601
|
NKG7
|
natural killer cell group 7 sequence
|
|
chr22_+_25398690
|
0.445
|
|
LOC388889
|
hypothetical LOC388889
|
|
chr14_-_24047261
|
0.435
|
NM_001836
|
CMA1
|
chymase 1, mast cell
|
|
chr5_+_61910318
|
0.422
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr6_-_153000226
|
0.419
|
NM_182961
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr4_+_54790190
|
0.414
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr18_-_59137488
|
0.413
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr6_+_50789215
|
0.412
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr11_-_110917632
|
0.403
|
|
|
|
|
chr1_-_79244948
|
0.385
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr5_-_88214720
|
0.382
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_127882192
|
0.375
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr2_-_175207521
|
0.362
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_+_26767657
|
0.361
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr21_-_27261276
|
0.355
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr20_+_2743613
|
0.344
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr7_+_129634935
|
0.323
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr5_+_140719773
|
0.317
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr2_-_220116062
|
0.312
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr5_+_140730014
|
0.309
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr17_-_40655347
|
0.302
|
|
LOC339192
|
hypothetical LOC339192
|
|
chr10_+_26767698
|
0.299
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr5_-_93472993
|
0.296
|
NM_001163417
NM_001163418
NM_032042
|
FAM172A
|
family with sequence similarity 172, member A
|
|
chr2_-_179623030
|
0.289
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr5_-_58370693
|
0.286
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr14_+_51397113
|
0.283
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr2_+_174968701
|
0.282
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr1_-_167603623
|
0.282
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr1_+_68070573
|
0.281
|
|
|
|
|
chr5_-_149515331
|
0.278
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr2_+_33514896
|
0.277
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr3_+_44571600
|
0.277
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr17_-_35097792
|
0.276
|
NM_033419
|
PGAP3
|
post-GPI attachment to proteins 3
|
|
chr18_+_7221136
|
0.276
|
NM_001105581
|
LRRC30
|
leucine rich repeat containing 30
|
|
chr6_-_89984214
|
0.276
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr2_+_39746525
|
0.274
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr4_-_10295412
|
0.273
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr6_+_29472310
|
0.271
|
NM_013936
|
OR12D2
|
olfactory receptor, family 12, subfamily D, member 2
|
|
chr8_-_49996353
|
0.270
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr3_+_120700057
|
0.269
|
NM_016589
|
C3orf1
|
chromosome 3 open reading frame 1
|
|
chr11_-_3104413
|
0.265
|
|
OSBPL5
|
oxysterol binding protein-like 5
|
|
chr2_+_169367345
|
0.265
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr9_+_107464558
|
0.264
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr17_-_34262862
|
0.260
|
|
RPL23
|
ribosomal protein L23
|
|
chr4_-_5071996
|
0.258
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr10_+_81882409
|
0.257
|
|
PLAC9
|
placenta-specific 9
|
|
chr17_+_35097851
|
0.255
|
NM_001005862
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr16_+_29581783
|
0.252
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr5_-_149515502
|
0.252
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr6_+_32003380
|
0.252
|
|
C2
|
complement component 2
|
|
chr2_-_213724577
|
0.248
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr17_+_40655060
|
0.245
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr2_+_174968751
|
0.245
|
|
SCRN3
|
secernin 3
|
|
chr9_+_134026836
|
0.244
|
|
NTNG2
|
netrin G2
|
|
chr14_+_19365447
|
0.244
|
NM_001004723
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2
|
|
chr5_+_40877053
|
0.243
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr16_-_85099941
|
0.243
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr10_-_96112609
|
0.242
|
NM_022451
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae)
|
|
chr9_-_90983501
|
0.240
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr12_+_8557402
|
0.240
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr18_+_21967813
|
0.237
|
NM_001025096
NM_001025097
NM_144662
|
PSMA8
|
proteasome (prosome, macropain) subunit, alpha type, 8
|
|
chr1_+_167604056
|
0.237
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr1_-_155788775
|
0.237
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr1_-_180822686
|
0.236
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr10_-_6144119
|
0.235
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr15_+_91244422
|
0.234
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr10_+_75174151
|
0.233
|
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chr5_+_140777454
|
0.233
|
NM_018927
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr10_+_75174125
|
0.231
|
NM_004922
NM_198597
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chr10_+_81882477
|
0.230
|
|
PLAC9
|
placenta-specific 9
|
|
chr6_+_126282078
|
0.226
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr11_-_74818685
|
0.224
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr10_+_115501202
|
0.223
|
NM_001193434
NM_001193435
NM_024889
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr15_+_91244563
|
0.222
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr12_-_6668680
|
0.222
|
|
ZNF384
|
zinc finger protein 384
|
|
chr1_+_167603817
|
0.222
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr12_+_12829807
|
0.221
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chrX_-_153632355
|
0.218
|
NM_001081573
NM_080612
|
GAB3
|
GRB2-associated binding protein 3
|
|
chr12_-_8584658
|
0.217
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr9_-_90983195
|
0.217
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr18_+_11841388
|
0.215
|
NM_020412
|
CHMP1B
|
chromatin modifying protein 1B
|
|
chr3_-_130600931
|
0.215
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr8_-_82186832
|
0.215
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr5_+_180514548
|
0.215
|
NM_206880
|
OR2V2
|
olfactory receptor, family 2, subfamily V, member 2
|
|
chr14_-_54727958
|
0.212
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr5_-_149515613
|
0.210
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr2_-_27469947
|
0.210
|
|
FTH1P3
|
ferritin, heavy polypeptide 1 pseudogene 3
|
|
chr2_-_200028977
|
0.209
|
|
SATB2
|
SATB homeobox 2
|
|
chr12_-_56157898
|
0.208
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr11_+_63412562
|
0.204
|
NM_017490
|
MARK2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr2_-_220116508
|
0.203
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr12_+_6950338
|
0.202
|
|
EMG1
|
EMG1 nucleolar protein homolog (S. cerevisiae)
|
|
chr12_-_6668880
|
0.202
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr6_+_28237529
|
0.201
|
NM_001145129
|
ZNF389
|
zinc finger protein 389
|
|
chr14_+_51397134
|
0.200
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr16_+_85169615
|
0.200
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr1_+_204797214
|
0.199
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr2_-_174968595
|
0.199
|
NM_004882
|
CIR1
|
corepressor interacting with RBPJ, 1
|
|
chr12_-_29828958
|
0.198
|
NM_175861
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr8_-_23768242
|
0.196
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr5_+_143171918
|
0.194
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr12_+_21570457
|
0.194
|
NM_030572
|
C12orf39
|
chromosome 12 open reading frame 39
|
|
chr17_+_55269974
|
0.193
|
|
|
|
|
chr14_+_51397059
|
0.192
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr14_+_51397166
|
0.191
|
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr11_-_74818858
|
0.191
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr8_-_78074851
|
0.190
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr6_-_34632061
|
0.190
|
NM_012391
|
SPDEF
|
SAM pointed domain containing ets transcription factor
|
|
chrX_-_110542040
|
0.187
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr3_+_25445071
|
0.187
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr2_+_102401580
|
0.187
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chrX_+_12795111
|
0.186
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr2_+_182029863
|
0.186
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr8_+_42247991
|
0.184
|
NM_001190720
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr11_+_22644732
|
0.183
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chrX_-_74292800
|
0.183
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chrX_-_31000079
|
0.182
|
NM_031894
|
FTHL17
|
ferritin, heavy polypeptide-like 17
|
|
chr6_-_41148061
|
0.182
|
NM_145063
|
C6orf130
|
chromosome 6 open reading frame 130
|
|
chr22_-_17892859
|
0.181
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr3_+_114733907
|
0.181
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr3_+_120495906
|
0.180
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr9_-_98368913
|
0.180
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr8_+_126054719
|
0.180
|
NM_152412
|
ZNF572
|
zinc finger protein 572
|
|
chr20_+_9406460
|
0.179
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr12_-_6950089
|
0.179
|
NM_007273
NM_001144831
|
PHB2
|
prohibitin 2
|
|
chr10_+_31650069
|
0.178
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr17_+_53587492
|
0.177
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chr21_-_34820916
|
0.176
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr4_-_156517394
|
0.176
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr2_+_31310485
|
0.176
|
|
EHD3
|
EH-domain containing 3
|
|
chr7_+_93388951
|
0.175
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr4_+_15080586
|
0.174
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr1_+_204797120
|
0.173
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr5_+_71538456
|
0.172
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_+_32003180
|
0.172
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr8_+_42247954
|
0.172
|
NM_001190722
NM_001556
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr11_+_46256161
|
0.171
|
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr6_-_116488458
|
0.171
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chrX_-_15312390
|
0.171
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr7_-_3049872
|
0.171
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr12_-_642786
|
0.171
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr11_-_9292581
|
0.170
|
|
TMEM41B
|
transmembrane protein 41B
|
|
chr14_-_44500840
|
0.170
|
|
KLHL28
|
kelch-like 28 (Drosophila)
|
|
chr17_+_4784524
|
0.169
|
|
RNF167
|
ring finger protein 167
|
|
chr11_-_60886038
|
0.169
|
NM_001161454
NM_153611
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr6_-_46997579
|
0.169
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr6_+_36961617
|
0.168
|
NM_152734
|
C6orf89
|
chromosome 6 open reading frame 89
|
|
chr4_-_156517423
|
0.168
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr3_+_148610516
|
0.168
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr20_+_19815075
|
0.168
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_+_12825818
|
0.168
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr14_-_56805118
|
0.167
|
|
|
|
|
chr17_-_43977273
|
0.166
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr12_-_109372462
|
0.165
|
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa
|
|
chr5_+_140767953
|
0.164
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr12_-_102968044
|
0.164
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr14_+_23008723
|
0.164
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr14_+_73073570
|
0.163
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr11_-_102219450
|
0.163
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr2_-_153282104
|
0.163
|
NM_017892
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae)
|
|
chr5_+_134012133
|
0.163
|
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chr2_+_97696462
|
0.163
|
NM_001079
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr13_+_25726755
|
0.162
|
NM_001260
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr4_-_101658094
|
0.161
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr17_+_11405798
|
0.161
|
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr6_+_45497796
|
0.161
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_-_111809929
|
0.159
|
NM_178175
|
LHFPL1
|
lipoma HMGIC fusion partner-like 1
|
|
chr1_-_53972464
|
0.158
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr17_-_64108843
|
0.158
|
|
FAM20A
|
family with sequence similarity 20, member A
|
|
chr20_+_33223434
|
0.158
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr13_-_42464301
|
0.156
|
NM_001002264
NM_033255
|
EPSTI1
|
epithelial stromal interaction 1 (breast)
|
|
chr4_+_86615275
|
0.155
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr7_-_91347915
|
0.155
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr12_+_6950204
|
0.154
|
NM_006331
|
EMG1
|
EMG1 nucleolar protein homolog (S. cerevisiae)
|
|
chr11_-_33847507
|
0.154
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr9_+_129226535
|
0.154
|
|
ZNF79
|
zinc finger protein 79
|
|
chr20_+_5679032
|
0.153
|
NM_152504
|
C20orf196
|
chromosome 20 open reading frame 196
|
|
chr1_+_204797079
|
0.153
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr8_+_42248008
|
0.152
|
NM_001190721
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
|
|
chr17_-_10362583
|
0.152
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr11_-_31789168
|
0.151
|
|
PAX6
|
paired box 6
|
|
chr22_+_35586950
|
0.151
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr8_-_13416554
|
0.151
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr2_+_165858586
|
0.151
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr16_+_67931178
|
0.150
|
|
NIP7
|
nuclear import 7 homolog (S. cerevisiae)
|
|
chr9_+_129226473
|
0.148
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr3_-_130362563
|
0.148
|
NM_020701
|
ISY1
|
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr6_-_116799548
|
0.148
|
|
LOC100287467
|
hypothetical LOC100287467
|
|
chr21_+_42697058
|
0.147
|
NM_001001895
NM_018961
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A
|
|
chr12_-_21985433
|
0.146
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr1_+_9926072
|
0.146
|
NM_022787
|
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1
|
|
chr5_+_140411144
|
0.145
|
NM_013340
|
PCDHB1
|
protocadherin beta 1
|
|
chr12_-_68369322
|
0.145
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr3_-_148607096
|
0.144
|
NM_032153
|
ZIC4
|
Zic family member 4
|
|
chr2_+_201099093
|
0.144
|
NM_001160033
NM_001160046
NM_152524
|
SGOL2
|
shugoshin-like 2 (S. pombe)
|
|
chr6_-_69401639
|
0.144
|
|
|
|
|
chr1_-_182273114
|
0.144
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr16_-_30992574
|
0.143
|
|
ZNF668
|
zinc finger protein 668
|