|
chr1_+_17719405
|
1.135
|
|
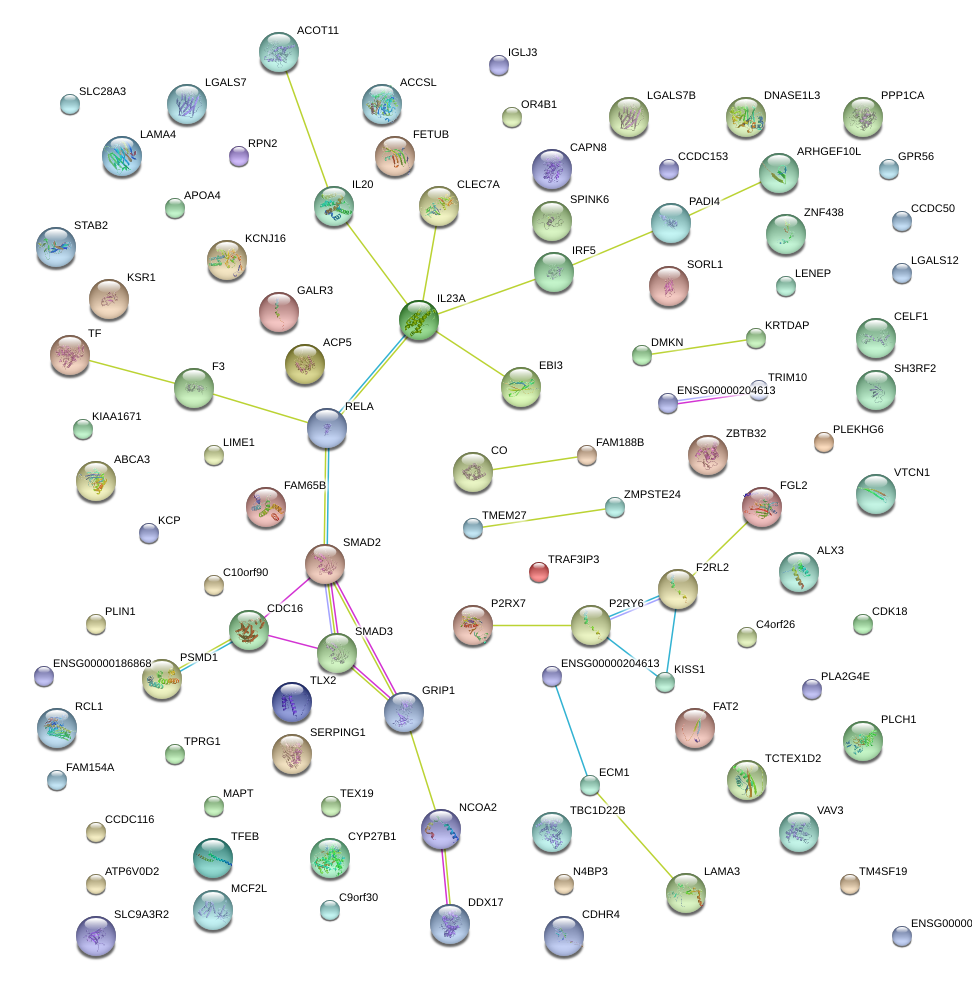
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr10_-_31328426
|
1.135
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr13_+_112746908
|
1.098
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_+_20899155
|
1.016
|
|
|
|
|
chr12_-_56445653
|
0.993
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr11_-_116199145
|
0.894
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr1_-_117554999
|
0.884
|
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr11_-_118571793
|
0.802
|
NM_001145018
|
CCDC153
|
coiled-coil domain containing 153
|
|
chr15_-_40089736
|
0.776
|
NM_001080490
|
PLA2G4E
|
phospholipase A2, group IVE
|
|
chr18_+_19706979
|
0.775
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr6_+_37360144
|
0.772
|
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr1_+_17507276
|
0.763
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr16_+_56219802
|
0.745
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr19_+_43971689
|
0.739
|
NM_001042507
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B
|
|
chr1_+_207995999
|
0.737
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr19_+_40895669
|
0.712
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr7_+_30927439
|
0.677
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr19_-_40684638
|
0.677
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr3_-_58175437
|
0.668
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr19_-_40673185
|
0.635
|
NM_207392
|
KRTDAP
|
keratinocyte differentiation-associated protein
|
|
chr10_-_128199986
|
0.621
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr15_+_65245546
|
0.604
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr1_-_43523825
|
0.596
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr7_+_128367961
|
0.580
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr19_+_4180494
|
0.579
|
NM_005755
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr1_-_202432165
|
0.566
|
NM_002256
|
KISS1
|
KiSS-1 metastasis-suppressor
|
|
chr3_+_190147633
|
0.560
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr17_+_41424665
|
0.556
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr22_+_21393107
|
0.552
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr12_+_6292077
|
0.522
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr1_+_205105776
|
0.513
|
NM_018724
|
IL20
|
interleukin 20
|
|
chr6_-_30236667
|
0.510
|
NM_006778
NM_052828
|
TRIM10
|
tripartite motif containing 10
|
|
chr22_+_23921072
|
0.505
|
|
KIAA1671
|
KIAA1671
|
|
chr5_+_177473071
|
0.504
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr17_+_65582981
|
0.494
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr22_+_21407066
|
0.491
|
|
|
|
|
chr17_+_77910411
|
0.487
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr22_+_20316968
|
0.486
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr11_+_48194937
|
0.484
|
NM_001005470
|
OR4B1
|
olfactory receptor, family 4, subfamily B, member 1
|
|
chr16_+_2023279
|
0.480
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr2_+_74595073
|
0.476
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr1_-_221919982
|
0.475
|
|
CAPN8
|
calpain 8
|
|
chr12_+_102505180
|
0.471
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chrX_-_15593074
|
0.468
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr22_+_36549334
|
0.466
|
NM_003614
|
GALR3
|
galanin receptor 3
|
|
chr1_+_40508336
|
0.464
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr17_+_22823151
|
0.461
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr6_-_41781655
|
0.458
|
|
TFEB
|
transcription factor EB
|
|
chr1_-_108032645
|
0.456
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr8_+_87180248
|
0.452
|
NM_152565
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2
|
|
chr22_+_21053972
|
0.446
|
|
|
|
|
chr9_+_102231616
|
0.444
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr1_-_221920023
|
0.431
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chr5_-_75954807
|
0.428
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr3_-_49812257
|
0.425
|
NM_001007540
|
CDHR4
|
cadherin-related family member 4
|
|
chr7_-_76667045
|
0.424
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr3_+_187840842
|
0.420
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr20_+_35266581
|
0.417
|
|
RPN2
|
ribophorin II
|
|
chr6_-_24985432
|
0.416
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr1_+_148747208
|
0.415
|
|
ECM1
|
extracellular matrix protein 1
|
|
chr9_-_86173232
|
0.405
|
NM_022127
|
SLC28A3
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 3
|
|
chr7_-_128330615
|
0.404
|
|
KCP
|
kielin/chordin-like protein
|
|
chr11_+_72660894
|
0.399
|
NM_004154
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr11_+_44026106
|
0.394
|
NM_001031854
|
ACCSL
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like
|
|
chr22_-_37214480
|
0.394
|
NM_001098505
NM_030881
|
DDX17
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
|
|
chr22_+_21116283
|
0.393
|
|
|
|
|
chr12_+_55018925
|
0.392
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr11_+_120966280
|
0.391
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr11_-_47502115
|
0.390
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr22_+_21484240
|
0.390
|
|
|
|
|
chr1_-_110414756
|
0.384
|
NM_006492
|
ALX3
|
ALX homeobox 3
|
|
chr4_+_76700281
|
0.377
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr11_+_57123927
|
0.376
|
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr5_+_145297490
|
0.375
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr5_+_147562531
|
0.375
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr1_+_153232685
|
0.373
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr1_-_178100204
|
0.373
|
|
|
|
|
chr15_-_88023576
|
0.369
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr11_-_65186790
|
0.369
|
|
RELA
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr12_-_10173947
|
0.367
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr1_+_203740306
|
0.365
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr12_-_65359019
|
0.363
|
|
GRIP1
|
glutamate receptor interacting protein 1
|
|
chr19_-_11549441
|
0.358
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_11549484
|
0.356
|
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr3_-_197549588
|
0.353
|
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr3_+_192529567
|
0.349
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr3_-_197549640
|
0.349
|
NM_138461
|
TM4SF19
|
transmembrane 4 L six family member 19
|
|
chr1_+_54780515
|
0.345
|
|
ACOT11
|
acyl-CoA thioesterase 11
|
|
chr9_+_4829763
|
0.340
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr3_+_134976965
|
0.338
|
|
TF
|
transferrin
|
|
chr3_-_156876798
|
0.338
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr2_-_85494639
|
0.337
|
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr3_+_185377341
|
0.337
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr9_+_5500544
|
0.336
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr1_+_160618143
|
0.335
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr22_+_21359175
|
0.335
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr15_+_38923914
|
0.333
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr21_+_29439768
|
0.331
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr12_-_68291208
|
0.331
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr1_+_151223180
|
0.330
|
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr19_+_40242634
|
0.328
|
|
HPN
|
hepsin
|
|
chr12_+_32546243
|
0.327
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_+_41712907
|
0.327
|
|
MDFI
|
MyoD family inhibitor
|
|
chr1_+_152644668
|
0.325
|
|
IL6R
|
interleukin 6 receptor
|
|
chr10_+_47867667
|
0.324
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr8_+_10567556
|
0.322
|
NM_001040032
|
C8orf74
|
chromosome 8 open reading frame 74
|
|
chr7_+_101978777
|
0.321
|
NM_001166339
NM_001031618
|
SPDYE2L
SPDYE2
|
WBSCR19-like protein 3
speedy homolog E2 (Xenopus laevis)
|
|
chr4_-_151990061
|
0.320
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr4_+_78172966
|
0.319
|
|
SEPT11
|
septin 11
|
|
chr1_+_148747110
|
0.318
|
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr17_-_70480385
|
0.317
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr8_+_22900863
|
0.316
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr9_-_139068269
|
0.316
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr1_-_36589744
|
0.315
|
|
|
|
|
chr7_-_98305599
|
0.313
|
NM_001134450
NM_001134451
NM_152913
|
TMEM130
|
transmembrane protein 130
|
|
chr1_-_26070330
|
0.312
|
NM_178422
|
PAQR7
|
progestin and adipoQ receptor family member VII
|
|
chr1_+_203740391
|
0.310
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr1_-_151804929
|
0.308
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr12_+_50981915
|
0.307
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr4_-_171161486
|
0.307
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr10_+_82268433
|
0.306
|
|
TSPAN14
|
tetraspanin 14
|
|
chr15_-_68781662
|
0.305
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_+_52325374
|
0.304
|
NM_015512
|
DNAH1
|
dynein, axonemal, heavy chain 1
|
|
chr7_+_26298136
|
0.303
|
|
SNX10
|
sorting nexin 10
|
|
chr1_+_110494654
|
0.301
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr19_-_43955984
|
0.297
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr8_+_22266256
|
0.297
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr16_-_30032330
|
0.295
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr7_-_98305551
|
0.295
|
|
TMEM130
|
transmembrane protein 130
|
|
chr3_+_125586241
|
0.291
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr21_-_42659671
|
0.290
|
NM_003225
|
TFF1
|
trefoil factor 1
|
|
chrX_-_10761729
|
0.290
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr13_-_59223074
|
0.289
|
|
|
|
|
chr12_+_6903886
|
0.288
|
NM_001007026
|
ATN1
|
atrophin 1
|
|
chr19_+_55123770
|
0.287
|
NM_012068
|
ATF5
|
activating transcription factor 5
|
|
chr11_-_62213946
|
0.286
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr5_+_176170041
|
0.285
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr14_-_93919305
|
0.283
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr20_+_42777419
|
0.283
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr15_+_39918302
|
0.279
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr11_-_64133908
|
0.276
|
|
|
|
|
chr13_-_99983945
|
0.274
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr3_-_11585264
|
0.272
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_+_185377430
|
0.268
|
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit
|
|
chr4_-_5879488
|
0.267
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr15_+_38923470
|
0.266
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr19_-_11549508
|
0.264
|
NM_001611
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_40939756
|
0.262
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr8_-_125649926
|
0.262
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr6_+_41118214
|
0.261
|
NM_001010873
NM_001159726
|
TSPO2
|
translocator protein 2
|
|
chr5_-_149446182
|
0.259
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr19_-_43926953
|
0.259
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr11_+_117903357
|
0.259
|
NM_001080441
|
TTC36
|
tetratricopeptide repeat domain 36
|
|
chr17_-_24069299
|
0.258
|
NM_001142624
NM_001142625
NM_001144943
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr8_-_25338472
|
0.257
|
NM_000825
NM_001083111
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone)
|
|
chr8_+_11599119
|
0.257
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr19_-_19599973
|
0.256
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr14_+_54291255
|
0.255
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr5_+_150000394
|
0.255
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr20_+_30480848
|
0.255
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chrX_+_24393264
|
0.254
|
NM_001142386
NM_005391
|
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3
|
|
chr1_-_6474346
|
0.254
|
NM_020631
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr8_-_11748330
|
0.254
|
|
|
|
|
chr10_-_49152918
|
0.253
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr17_-_38421363
|
0.252
|
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr21_-_34908614
|
0.252
|
NM_203417
|
RCAN1
|
regulator of calcineurin 1
|
|
chr1_-_7998265
|
0.251
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr2_+_218833982
|
0.250
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr8_+_120289735
|
0.250
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr6_-_112001590
|
0.250
|
NM_001164282
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr20_+_42777298
|
0.249
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr14_+_76362467
|
0.249
|
NM_194287
|
C14orf166B
|
chromosome 14 open reading frame 166B
|
|
chr10_+_35575958
|
0.248
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr22_+_21340757
|
0.247
|
|
|
|
|
chr1_-_151333624
|
0.246
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr5_+_175241449
|
0.245
|
|
CPLX2
|
complexin 2
|
|
chr8_-_143820825
|
0.244
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr3_+_153074118
|
0.243
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr10_-_46601687
|
0.243
|
NM_001137556
NM_001137549
NM_001137548
|
FAM25B
FAM25G
FAM25C
|
family with sequence similarity 25, member B
family with sequence similarity 25, member G
family with sequence similarity 25, member C
|
|
chr19_-_6621078
|
0.242
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr21_-_36760594
|
0.242
|
NM_012130
|
CLDN14
|
claudin 14
|
|
chr10_+_88770025
|
0.242
|
NM_001146157
|
FAM25A
|
family with sequence similarity 25, member A
|
|
chr5_-_141041982
|
0.242
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr15_+_57691382
|
0.242
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr17_+_23824784
|
0.241
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr5_-_141041933
|
0.239
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr1_-_92725017
|
0.239
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr15_+_88696480
|
0.239
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr17_+_35718948
|
0.238
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr4_+_661710
|
0.238
|
NM_002477
|
MYL5
|
myosin, light chain 5, regulatory
|
|
chr5_-_150501393
|
0.238
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr5_-_86739683
|
0.237
|
|
CCNH
|
cyclin H
|
|
chr12_-_47545816
|
0.236
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chr4_-_78038025
|
0.236
|
NM_001029870
|
ANKRD56
|
ankyrin repeat domain 56
|
|
chr17_+_7225088
|
0.235
|
NM_003985
|
TNK1
|
tyrosine kinase, non-receptor, 1
|
|
chr19_+_46489987
|
0.235
|
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr1_-_202387532
|
0.234
|
|
ETNK2
|
ethanolamine kinase 2
|
|
chr19_-_63177713
|
0.234
|
NM_152474
|
C19orf18
|
chromosome 19 open reading frame 18
|
|
chr5_-_131658838
|
0.233
|
|
|
|
|
chrX_+_47114925
|
0.233
|
NM_003446
|
ZNF157
|
zinc finger protein 157
|