|
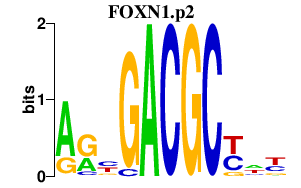
chr14_+_36196523
|
2.058
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr2_-_142605010
|
1.764
|
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr10_+_103103797
|
1.657
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr2_-_54051290
|
1.417
|
NM_014614
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4
|
|
chr17_+_40581704
|
1.221
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr20_-_2592777
|
1.126
|
NM_006899
NM_174855
NM_174856
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr20_+_29656744
|
1.099
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr20_-_25010756
|
1.083
|
NM_014588
NM_199425
|
VSX1
|
visual system homeobox 1
|
|
chr14_+_30564433
|
1.079
|
NM_001128126
NM_007077
|
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit
|
|
chr1_-_45924770
|
1.070
|
NM_021639
|
GPBP1L1
|
GC-rich promoter binding protein 1-like 1
|
|
chr20_+_3749164
|
1.052
|
NM_018347
|
C20orf29
|
chromosome 20 open reading frame 29
|
|
chrX_+_90920915
|
1.038
|
NM_032967
|
PCDH11X
PCDH11Y
|
protocadherin 11 X-linked
protocadherin 11 Y-linked
|
|
chr4_-_170770111
|
1.037
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr3_+_139388770
|
0.943
|
NM_014154
NM_015396
NM_213654
|
ARMC8
|
armadillo repeat containing 8
|
|
chr14_-_87859284
|
0.933
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr4_+_81475897
|
0.915
|
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chr17_-_42250961
|
0.878
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr2_+_30523586
|
0.870
|
NM_182551
NM_001002257
|
LCLAT1
|
lysocardiolipin acyltransferase 1
|
|
chr20_+_3775421
|
0.869
|
NM_020746
|
MAVS
|
mitochondrial antiviral signaling protein
|
|
chr5_+_80292213
|
0.837
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr20_-_48163071
|
0.821
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr14_+_34585098
|
0.810
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr17_+_11442472
|
0.797
|
NM_001372
|
DNAH9
|
dynein, axonemal, heavy chain 9
|
|
chr12_+_56230073
|
0.776
|
|
KIF5A
|
kinesin family member 5A
|
|
chr10_+_112394094
|
0.771
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr3_-_191321367
|
0.769
|
|
LEPREL1
|
leprecan-like 1
|
|
chr8_+_65656164
|
0.767
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr15_-_32875040
|
0.759
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr1_+_82038669
|
0.750
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr9_+_8848130
|
0.749
|
|
|
|
|
chr5_-_100266769
|
0.747
|
NM_175052
NM_005668
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr16_+_25610847
|
0.747
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr12_+_56230047
|
0.735
|
|
KIF5A
|
kinesin family member 5A
|
|
chr12_+_56230110
|
0.734
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr1_-_183210155
|
0.730
|
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr19_+_2240996
|
0.720
|
|
|
|
|
chr2_+_120018494
|
0.719
|
NM_001029996
|
PCDP1
|
primary ciliary dyskinesia protein 1
|
|
chr20_-_19981005
|
0.715
|
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr14_-_22127902
|
0.714
|
|
DAD1
|
defender against cell death 1
|
|
chr16_-_56038771
|
0.702
|
NM_020313
|
CIAPIN1
|
cytokine induced apoptosis inhibitor 1
|
|
chr14_-_91372365
|
0.690
|
NM_001128595
NM_152332
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr16_+_56038837
|
0.689
|
NM_020312
|
COQ9
|
coenzyme Q9 homolog (S. cerevisiae)
|
|
chr22_+_47263919
|
0.687
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr11_-_8982107
|
0.678
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr15_+_39008822
|
0.670
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr1_-_25129354
|
0.666
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr17_+_33761234
|
0.664
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr22_+_27798887
|
0.661
|
NM_001039570
NM_032045
|
KREMEN1
|
kringle containing transmembrane protein 1
|
|
chr15_-_47235057
|
0.660
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr2_-_200424140
|
0.656
|
|
LOC348751
|
hypothetical LOC348751
|
|
chr1_+_95355415
|
0.645
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr11_-_75599433
|
0.645
|
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr5_+_80292313
|
0.643
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr11_-_107969554
|
0.639
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr17_+_76133206
|
0.637
|
NM_001163034
NM_020761
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr17_-_35273947
|
0.636
|
NM_012481
NM_183228
NM_183229
NM_183230
NM_183231
NM_183232
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr5_-_134899537
|
0.633
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr16_+_1143738
|
0.630
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr17_+_76133337
|
0.629
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr11_-_132318741
|
0.625
|
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chrX_-_21586337
|
0.623
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr2_+_187059115
|
0.618
|
NM_018471
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr8_+_21937533
|
0.614
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr4_-_21559471
|
0.613
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr12_-_79855580
|
0.604
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr14_-_22127954
|
0.604
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chrX_-_137621060
|
0.600
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr4_+_3738093
|
0.595
|
NM_000683
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr11_-_27341251
|
0.593
|
NM_030771
NM_080654
|
CCDC34
|
coiled-coil domain containing 34
|
|
chr1_+_78243181
|
0.593
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr2_-_219614481
|
0.586
|
NM_152389
NM_194302
|
CCDC108
|
coiled-coil domain containing 108
|
|
chr12_+_4889309
|
0.582
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr19_-_11452439
|
0.575
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr2_+_190014403
|
0.573
|
NM_032168
|
WDR75
|
WD repeat domain 75
|
|
chr8_-_38245694
|
0.573
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr20_+_254205
|
0.571
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr19_-_51856129
|
0.571
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr11_-_93774223
|
0.567
|
NM_016540
|
GPR83
|
G protein-coupled receptor 83
|
|
chr2_+_232534535
|
0.566
|
NM_152383
|
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2
|
|
chr17_-_37586716
|
0.566
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr2_+_187059277
|
0.566
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr8_+_87424074
|
0.552
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr10_+_124211353
|
0.552
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr17_+_7125702
|
0.549
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr2_-_43676603
|
0.548
|
NM_001083953
NM_022065
|
THADA
|
thyroid adenoma associated
|
|
chr2_+_190014432
|
0.544
|
|
WDR75
|
WD repeat domain 75
|
|
chr17_-_17081217
|
0.537
|
NM_144606
NM_144997
|
FLCN
|
folliculin
|
|
chr20_-_18395622
|
0.536
|
NM_001099407
|
C20orf12
|
chromosome 20 open reading frame 12
|
|
chr11_-_88864061
|
0.533
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr15_+_47235243
|
0.523
|
NM_001001556
|
GALK2
|
galactokinase 2
|
|
chr2_+_99320181
|
0.520
|
NM_015904
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr17_-_46553083
|
0.517
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr17_+_34871775
|
0.513
|
|
CDK12
|
cyclin-dependent kinase 12
|
|
chr3_+_3816079
|
0.512
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr2_-_202215593
|
0.511
|
NM_152388
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chrX_+_101267315
|
0.511
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr4_-_111763655
|
0.511
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr15_+_63948823
|
0.504
|
NM_004663
|
RAB11A
|
RAB11A, member RAS oncogene family
|
|
chr1_-_25129305
|
0.503
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr10_+_101282679
|
0.499
|
NM_145285
|
NKX2-3
|
NK2 transcription factor related, locus 3 (Drosophila)
|
|
chr3_-_121944052
|
0.496
|
NM_173825
|
RABL3
|
RAB, member of RAS oncogene family-like 3
|
|
chr2_+_187059237
|
0.494
|
|
ZC3H15
|
zinc finger CCCH-type containing 15
|
|
chr3_+_121944247
|
0.482
|
NM_005513
|
GTF2E1
|
general transcription factor IIE, polypeptide 1, alpha 56kDa
|
|
chr17_-_45562112
|
0.478
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr14_+_51526071
|
0.475
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr2_+_220007943
|
0.473
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr14_+_92330383
|
0.472
|
NM_005113
|
GOLGA5
|
golgin A5
|
|
chr9_+_96176638
|
0.467
|
NM_032558
|
HIATL1
|
hippocampus abundant transcript-like 1
|
|
chr2_+_68548173
|
0.464
|
NM_173545
|
APLF
|
aprataxin and PNKP like factor
|
|
chr13_-_52320774
|
0.460
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr2_-_202216451
|
0.457
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr4_+_172971149
|
0.457
|
NM_001034845
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6
|
|
chr18_-_30056431
|
0.456
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr4_-_155631861
|
0.448
|
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr4_+_119419354
|
0.448
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr12_-_79855618
|
0.446
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr11_+_7997366
|
0.445
|
|
TUB
|
tubby homolog (mouse)
|
|
chr11_+_65311104
|
0.445
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr22_+_40195058
|
0.441
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr2_-_176656935
|
0.440
|
NM_001080458
|
EVX2
|
even-skipped homeobox 2
|
|
chr3_-_134863316
|
0.437
|
NM_007027
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1
|
|
chr2_+_210575600
|
0.437
|
NM_199229
|
RPE
|
ribulose-5-phosphate-3-epimerase
|
|
chr2_-_154043403
|
0.437
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr10_-_69762654
|
0.433
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr8_+_121892719
|
0.429
|
|
|
|
|
chr14_+_64077171
|
0.427
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr16_-_730889
|
0.426
|
NM_022493
|
NARFL
|
nuclear prelamin A recognition factor-like
|
|
chr2_+_185171480
|
0.426
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr5_-_134397740
|
0.423
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr3_-_192063158
|
0.421
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr15_-_38000379
|
0.418
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr3_-_121944040
|
0.416
|
|
|
|
|
chr5_+_93980108
|
0.412
|
NM_032290
|
ANKRD32
|
ankyrin repeat domain 32
|
|
chr14_+_51525942
|
0.412
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr2_+_210575566
|
0.408
|
NM_006916
|
RPE
|
ribulose-5-phosphate-3-epimerase
|
|
chr20_-_35589496
|
0.408
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr15_-_76210611
|
0.405
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr2_-_20114761
|
0.405
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr4_-_1683712
|
0.405
|
NM_006527
|
SLBP
|
stem-loop binding protein
|
|
chr19_-_16599986
|
0.404
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr15_+_76343540
|
0.402
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr10_-_119124727
|
0.400
|
NM_173791
|
PDZD8
|
PDZ domain containing 8
|
|
chr7_-_50828159
|
0.398
|
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr7_-_33068699
|
0.397
|
NM_001002010
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr2_-_100400481
|
0.396
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr11_+_65311142
|
0.393
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr16_+_84490267
|
0.393
|
NM_002163
|
IRF8
|
interferon regulatory factor 8
|
|
chr14_-_20642214
|
0.390
|
|
|
|
|
chr17_-_38229790
|
0.389
|
NM_003766
|
BECN1
|
beclin 1, autophagy related
|
|
chr10_-_6059460
|
0.387
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr2_+_223433919
|
0.384
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr17_-_44663107
|
0.384
|
NM_001143804
NM_178500
|
PHOSPHO1
|
phosphatase, orphan 1
|
|
chr17_-_71287417
|
0.382
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr17_+_77478858
|
0.382
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr10_-_127574957
|
0.379
|
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr9_-_95757365
|
0.379
|
NM_021570
|
BARX1
|
BARX homeobox 1
|
|
chr17_+_40581617
|
0.377
|
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chrX_+_105856531
|
0.377
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr5_-_45731976
|
0.374
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr2_-_38683119
|
0.371
|
|
HNRPLL
|
heterogeneous nuclear ribonucleoprotein L-like
|
|
chr12_+_87060176
|
0.368
|
NM_181783
|
TMTC3
|
transmembrane and tetratricopeptide repeat containing 3
|
|
chr20_-_1395463
|
0.368
|
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chr6_+_99389300
|
0.368
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr2_+_73466329
|
0.367
|
NM_015120
|
ALMS1
|
Alstrom syndrome 1
|
|
chr16_+_70486933
|
0.366
|
NM_014761
|
KIAA0174
|
KIAA0174
|
|
chr13_-_72253970
|
0.366
|
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr2_-_196641682
|
0.364
|
NM_018897
|
DNAH7
|
dynein, axonemal, heavy chain 7
|
|
chr17_-_13445942
|
0.359
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr5_+_112340326
|
0.357
|
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chrX_-_17789276
|
0.356
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr15_-_37999717
|
0.356
|
|
GPR176
|
G protein-coupled receptor 176
|
|
chr9_+_134895882
|
0.356
|
NM_001122823
NM_012087
|
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa
|
|
chr13_+_44461657
|
0.355
|
NM_018559
|
KIAA1704
|
KIAA1704
|
|
chr17_+_43165608
|
0.354
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr16_+_69115227
|
0.353
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr4_-_66218777
|
0.353
|
|
EPHA5
|
EPH receptor A5
|
|
chr12_+_68923477
|
0.351
|
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr12_-_29425356
|
0.351
|
NM_016570
|
ERGIC2
|
ERGIC and golgi 2
|
|
chr16_+_69115225
|
0.348
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr6_+_117305068
|
0.348
|
NM_173560
|
RFX6
|
regulatory factor X, 6
|
|
chr9_-_95757448
|
0.348
|
|
BARX1
|
BARX homeobox 1
|
|
chr13_+_44592585
|
0.348
|
NM_004128
|
GTF2F2
|
general transcription factor IIF, polypeptide 2, 30kDa
|
|
chr1_-_209818342
|
0.348
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr8_+_32525269
|
0.347
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr20_+_56660212
|
0.347
|
|
STX16
|
syntaxin 16
|
|
chr6_+_1335067
|
0.347
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr14_+_31616196
|
0.345
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr6_+_117693413
|
0.344
|
NM_153453
NM_182645
|
VGLL2
|
vestigial like 2 (Drosophila)
|
|
chr19_+_40325993
|
0.343
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chr16_+_70486900
|
0.341
|
|
KIAA0174
|
KIAA0174
|
|
chr18_-_11138760
|
0.340
|
NM_022068
|
FAM38B
|
family with sequence similarity 38, member B
|
|
chr12_+_56406296
|
0.339
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr2_+_170044194
|
0.338
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chr5_+_88220846
|
0.338
|
|
|
|
|
chr14_-_49122813
|
0.337
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr13_-_72254071
|
0.334
|
NM_001128226
NM_014953
|
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr4_+_57066131
|
0.334
|
NM_206919
|
ARL9
|
ADP-ribosylation factor-like 9
|
|
chr3_+_50687675
|
0.332
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr14_-_37134191
|
0.331
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr6_-_90405156
|
0.330
|
NM_020466
|
LYRM2
|
LYR motif containing 2
|