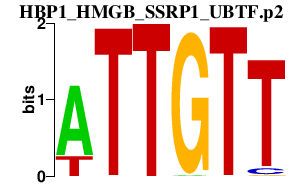
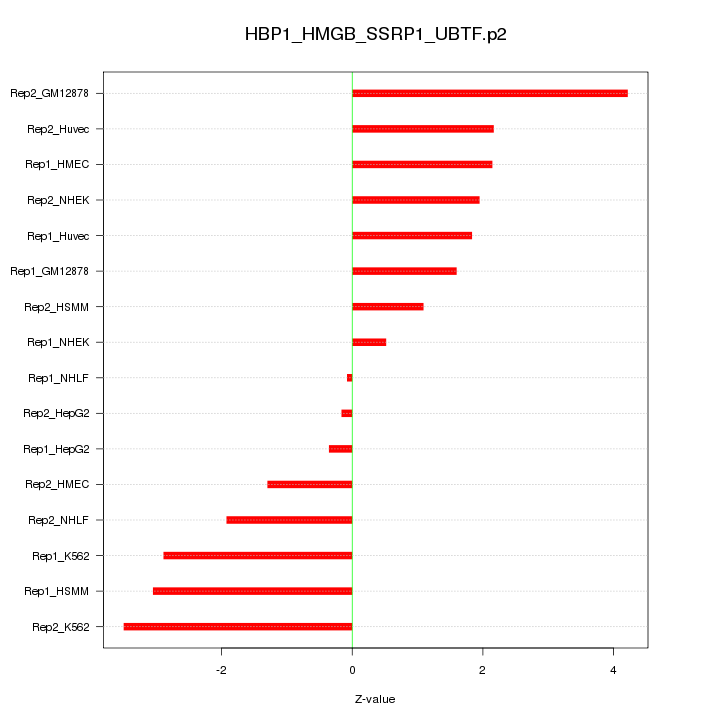
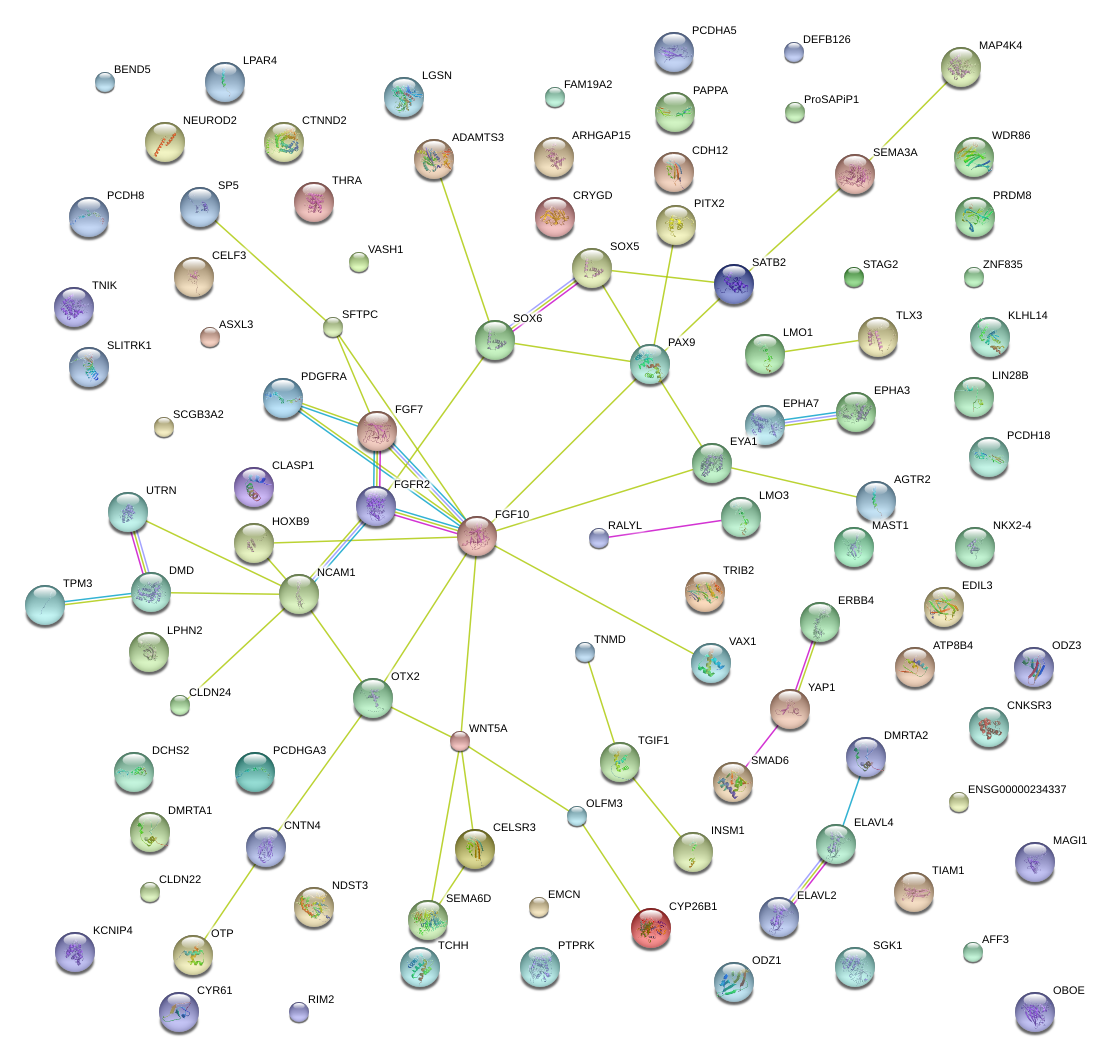
|
chr6_+_105511615
|
7.221
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr14_-_56342097
|
5.730
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr12_-_16649211
|
5.665
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr13_-_52320774
|
5.254
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr11_-_16380967
|
5.068
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_-_72436519
|
4.811
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr5_+_170668892
|
4.438
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr14_+_36196523
|
4.071
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr2_-_200033445
|
3.998
|
|
SATB2
|
SATB homeobox 2
|
|
chr5_-_88004891
|
3.938
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr18_+_3441589
|
3.641
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr4_-_101658094
|
3.465
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr5_-_44424423
|
3.426
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr9_-_23816062
|
3.389
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr13_+_99431417
|
3.246
|
|
|
|
|
chr15_-_48198697
|
3.045
|
NM_024837
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr2_+_12775814
|
3.003
|
|
|
|
|
chr6_-_128883125
|
2.916
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr1_-_102234959
|
2.798
|
|
OLFM3
|
olfactomedin 3
|
|
chr18_-_28606839
|
2.707
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr15_+_64781723
|
2.639
|
NM_005585
|
SMAD6
|
SMAD family member 6
|
|
chr8_-_72437020
|
2.615
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr15_+_47502666
|
2.586
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr3_+_2117246
|
2.520
|
NM_175607
|
CNTN4
|
contactin 4
|
|
chr12_-_16652202
|
2.477
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_-_20594645
|
2.379
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr2_-_208697441
|
2.350
|
NM_006891
|
CRYGD
|
crystallin, gamma D
|
|
chrX_+_77889861
|
2.302
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr17_-_44058612
|
2.246
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chrX_+_99726445
|
2.219
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr1_-_150353179
|
2.167
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr6_-_94185780
|
2.167
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr3_-_48675321
|
2.163
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr1_+_81544432
|
2.118
|
|
LPHN2
|
latrophilin 2
|
|
chr8_-_72436832
|
2.060
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_+_54790190
|
2.047
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr3_+_89239494
|
1.985
|
|
EPHA3
|
EPH receptor A3
|
|
chr17_-_35017650
|
1.983
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr2_+_171280046
|
1.938
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr1_+_85819004
|
1.924
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chrX_+_115215985
|
1.880
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr18_-_29412148
|
1.860
|
|
|
|
|
chr20_+_20296744
|
1.853
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr11_+_112337199
|
1.837
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr2_-_100088691
|
1.824
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr2_-_213111525
|
1.820
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr20_-_3097070
|
1.806
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr13_-_83354474
|
1.795
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr15_+_45797977
|
1.794
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr3_-_55496370
|
1.793
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr1_-_50661716
|
1.776
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr4_+_119174947
|
1.771
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr5_+_140181405
|
1.737
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr7_-_83662152
|
1.714
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr14_+_76298693
|
1.687
|
|
VASH1
|
vasohibin 1
|
|
chr12_-_16650697
|
1.668
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_123062143
|
1.655
|
|
STAG2
|
stromal antigen 2
|
|
chr12_-_16650687
|
1.638
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_-_111777651
|
1.629
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr10_-_118887801
|
1.607
|
NM_001112704
NM_199131
|
VAX1
|
ventral anterior homeobox 1
|
|
chr2_-_72228427
|
1.606
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr20_-_21326046
|
1.605
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr1_-_49014993
|
1.597
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr5_+_140777454
|
1.584
|
NM_018927
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr8_+_85258257
|
1.580
|
|
RALYL
|
RALY RNA binding protein-like
|
|
chr5_-_11956964
|
1.567
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr4_-_138673078
|
1.567
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr3_+_89239363
|
1.564
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr19_+_12810258
|
1.562
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr2_+_101680594
|
1.559
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr4_-_73653331
|
1.554
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr2_+_12775336
|
1.552
|
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr19_-_61876057
|
1.551
|
NM_001005850
|
ZNF835
|
zinc finger protein 835
|
|
chr3_-_65999548
|
1.521
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr10_-_123346148
|
1.520
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_+_143603439
|
1.511
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr17_+_35471971
|
1.500
|
NM_001190919
|
THRA
|
thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian)
|
|
chr13_-_83354425
|
1.496
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr21_-_31853160
|
1.491
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr20_+_71230
|
1.485
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr2_-_100088760
|
1.485
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr18_+_29412538
|
1.483
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr4_-_184480572
|
1.472
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr2_+_149349643
|
1.467
|
|
|
|
|
chr1_-_152431175
|
1.465
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr5_-_76970264
|
1.462
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr5_+_147238466
|
1.445
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr1_-_149955870
|
1.444
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr7_-_150737980
|
1.438
|
NM_198285
|
WDR86
|
WD repeat domain 86
|
|
chrX_-_33267646
|
1.430
|
NM_000109
|
DMD
|
dystrophin
|
|
chr4_-_155632317
|
1.426
|
NM_001142552
NM_001142553
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr8_+_104900591
|
1.397
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr6_-_134680888
|
1.393
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr5_-_22889487
|
1.392
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr9_+_117955890
|
1.383
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr3_+_148593911
|
1.374
|
|
|
|
|
chr4_+_183302105
|
1.367
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr12_-_23995109
|
1.363
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr6_-_64087806
|
1.356
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr4_-_155631861
|
1.351
|
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr11_+_101488380
|
1.349
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr5_-_83715947
|
1.344
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr4_+_81337663
|
1.336
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr19_+_12995782
|
1.323
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr14_-_36058648
|
1.315
|
NM_003317
|
NKX2-1
|
NK2 homeobox 1
|
|
chr18_-_30056431
|
1.313
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr10_+_11246934
|
1.312
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr6_-_134540666
|
1.306
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr13_-_98428244
|
1.296
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr10_+_70517823
|
1.270
|
NM_002727
|
SRGN
|
serglycin
|
|
chr5_+_15553736
|
1.266
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr1_-_155095289
|
1.264
|
NM_014215
|
INSRR
|
insulin receptor-related receptor
|
|
chr9_-_16717887
|
1.263
|
|
BNC2
|
basonuclin 2
|
|
chr9_-_14304036
|
1.262
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr5_+_98132898
|
1.247
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr12_+_83954229
|
1.246
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr19_+_11510578
|
1.244
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr2_-_162808123
|
1.237
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr12_-_8584658
|
1.233
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr10_+_70517863
|
1.231
|
|
SRGN
|
serglycin
|
|
chr8_+_85258007
|
1.224
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr5_-_83716346
|
1.222
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chrX_+_28515479
|
1.211
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr3_-_55496607
|
1.210
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr10_+_111957292
|
1.210
|
NM_130439
|
MXI1
|
MAX interactor 1
|
|
chr2_-_165768798
|
1.201
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr11_-_31789431
|
1.200
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr15_+_58083712
|
1.197
|
NM_012182
|
FOXB1
|
forkhead box B1
|
|
chr13_+_96592051
|
1.197
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_+_28418668
|
1.195
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_-_31789334
|
1.189
|
|
PAX6
|
paired box 6
|
|
chr14_+_76297885
|
1.185
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr9_-_94338072
|
1.184
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr11_-_31789360
|
1.176
|
|
PAX6
|
paired box 6
|
|
chr8_+_85258056
|
1.163
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr12_-_83954185
|
1.158
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr4_+_86918882
|
1.157
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_238842071
|
1.156
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr13_+_107753764
|
1.152
|
|
|
|
|
chr1_-_214963279
|
1.146
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr11_-_31789168
|
1.145
|
|
PAX6
|
paired box 6
|
|
chr12_-_23995220
|
1.145
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_+_104837004
|
1.145
|
|
|
|
|
chrX_-_131401319
|
1.143
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr12_-_60872814
|
1.142
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr1_-_215377718
|
1.141
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr13_+_51334117
|
1.140
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chrX_+_152562013
|
1.137
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr2_+_143603368
|
1.137
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr15_+_74416156
|
1.135
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr8_+_70567634
|
1.133
|
|
SULF1
|
sulfatase 1
|
|
chr4_+_52612197
|
1.132
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr4_-_64957595
|
1.132
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr11_-_114880275
|
1.131
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr11_+_131285747
|
1.124
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr11_-_20138445
|
1.122
|
NM_001029865
|
DBX1
|
developing brain homeobox 1
|
|
chr14_-_59167193
|
1.122
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr2_+_185171682
|
1.119
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr17_+_56843893
|
1.114
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr12_-_88270375
|
1.113
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr9_+_126460511
|
1.109
|
|
|
|
|
chr8_-_87824996
|
1.105
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr11_+_131286513
|
1.102
|
|
NTM
|
neurotrimin
|
|
chr7_+_113842284
|
1.102
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chrX_-_10605726
|
1.095
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_-_142551587
|
1.094
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr12_-_7487989
|
1.084
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr6_+_50894397
|
1.082
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr5_-_76970129
|
1.081
|
|
OTP
|
orthopedia homeobox
|
|
chr4_+_170817787
|
1.067
|
|
CLCN3
|
chloride channel 3
|
|
chr11_+_22316242
|
1.060
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr2_+_171280182
|
1.060
|
|
|
|
|
chr5_-_22889192
|
1.056
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr5_-_147142457
|
1.056
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr8_+_102574012
|
1.051
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_219119430
|
1.050
|
|
HLX
|
H2.0-like homeobox
|
|
chr17_+_71095733
|
1.049
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr5_-_134899537
|
1.038
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr17_-_35017691
|
1.036
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr13_-_78075683
|
1.033
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr7_-_11838260
|
1.032
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr3_+_182912405
|
1.032
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr7_+_28415495
|
1.031
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr1_-_102235377
|
1.029
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chrX_-_116991728
|
1.025
|
NM_033495
NM_001168300
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr12_-_98072602
|
1.019
|
NM_020140
NM_181670
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr7_+_18501876
|
1.018
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr14_+_36200867
|
1.016
|
|
PAX9
|
paired box 9
|
|
chrX_-_138114100
|
1.015
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr9_+_102830851
|
1.014
|
NM_207299
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr4_-_111777584
|
1.011
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr13_-_37341859
|
1.009
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr11_-_102219450
|
1.007
|
NM_002422
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase)
|
|
chr7_-_19123765
|
1.007
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr17_-_76985538
|
1.000
|
|
|
|
|
chr9_+_27099138
|
0.988
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_+_154373323
|
0.985
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr3_+_116824840
|
0.983
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr4_-_165523856
|
0.981
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr8_-_114518194
|
0.979
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|