|
chr10_-_99521701
|
7.279
|
NM_003015
|
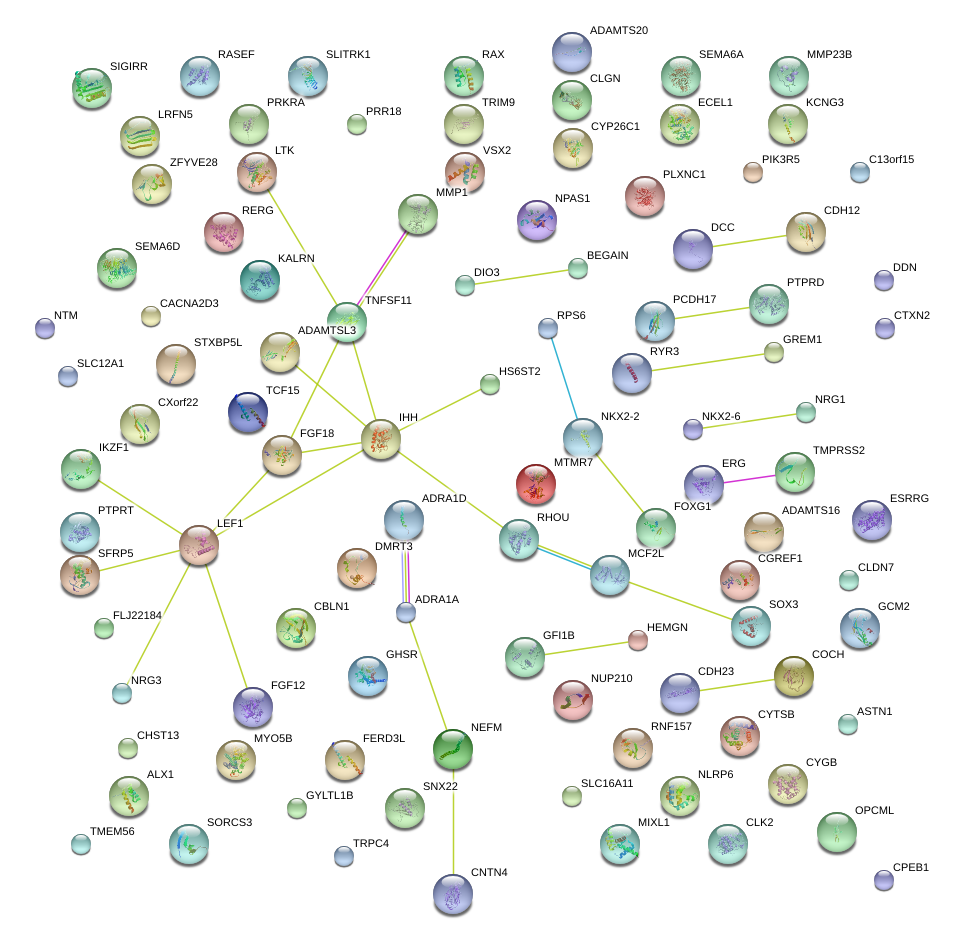
SFRP5
|
secreted frizzled-related protein 5
|
|
chr3_-_173648896
|
4.036
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chr7_-_19151509
|
3.961
|
NM_152898
|
FERD3L
|
Fer3-like (Drosophila)
|
|
chr21_-_38954433
|
3.853
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_+_83625049
|
3.587
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr14_+_28306633
|
3.582
|
|
FOXG1
|
forkhead box G1
|
|
chr4_-_2390050
|
3.542
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chrX_-_139414890
|
3.236
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr2_-_42573980
|
3.235
|
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr20_-_21442663
|
3.214
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr20_-_538909
|
3.141
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr18_-_55091596
|
3.052
|
NM_013435
|
RAX
|
retina and anterior neural fold homeobox
|
|
chr8_-_26780665
|
2.806
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr10_+_94811010
|
2.792
|
NM_183374
|
CYP26C1
|
cytochrome P450, family 26, subfamily C, polypeptide 1
|
|
chrX_-_131918905
|
2.769
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr3_+_54131613
|
2.764
|
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr8_+_31616809
|
2.698
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr14_+_30413393
|
2.672
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr16_-_85099941
|
2.641
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr8_+_24828359
|
2.626
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr13_+_42046182
|
2.614
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr11_+_268569
|
2.609
|
NM_138329
|
NLRP6
|
NLR family, pyrin domain containing 6
|
|
chr8_-_26780645
|
2.601
|
|
|
|
|
chr2_-_219633481
|
2.501
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr15_-_39593285
|
2.494
|
NM_001135685
NM_002344
NM_206961
|
LTK
|
leukocyte receptor tyrosine kinase
|
|
chr1_-_175400441
|
2.485
|
|
ASTN1
|
astrotactin 1
|
|
chr17_-_6887965
|
2.452
|
NM_153357
|
SLC16A11
|
solute carrier family 16, member 11 (monocarboxylic acid transporter 11)
|
|
chr17_-_72045218
|
2.429
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr8_-_24828134
|
2.403
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr14_-_100104014
|
2.394
|
NM_001159531
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr15_+_30797466
|
2.366
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr4_-_109307325
|
2.343
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr11_+_131286513
|
2.264
|
|
NTM
|
neurotrimin
|
|
chr13_-_37341859
|
2.259
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr8_-_17315386
|
2.254
|
NM_004686
|
MTMR7
|
myotubularin related protein 7
|
|
chr3_+_125786195
|
2.238
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr18_-_45975163
|
2.212
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr3_+_122109716
|
2.204
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr21_-_41801778
|
2.188
|
NM_001135099
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr4_-_141568210
|
2.169
|
NM_001130675
NM_004362
|
CLGN
|
calmegin
|
|
chr12_+_84198015
|
2.147
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr20_-_41251878
|
2.145
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr5_-_88004891
|
2.144
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr3_+_2117246
|
2.107
|
NM_175607
|
CNTN4
|
contactin 4
|
|
chr14_+_101097440
|
2.095
|
NM_001362
|
DIO3
|
deiodinase, iodothyronine, type III
|
|
chr21_-_41801946
|
2.072
|
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr7_+_50314801
|
2.064
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_-_215377718
|
2.062
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr15_+_45263517
|
2.045
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr21_-_41801950
|
2.034
|
NM_005656
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr14_-_50631407
|
2.033
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr1_+_224477941
|
2.001
|
NM_031944
|
MIXL1
|
Mix1 homeobox-like 1 (Xenopus laevis)
|
|
chr6_-_10990048
|
1.976
|
NM_004752
|
GCM2
|
glial cells missing homolog 2 (Drosophila)
|
|
chr13_+_57105859
|
1.974
|
|
PCDH17
|
protocadherin 17
|
|
chr3_+_54131731
|
1.960
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr5_-_22248630
|
1.919
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr12_-_15265490
|
1.915
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr8_-_23619866
|
1.914
|
NM_001136271
|
NKX2-6
|
NK2 transcription factor related, locus 6 (Drosophila)
|
|
chr8_-_17315171
|
1.907
|
|
MTMR7
|
myotubularin related protein 7
|
|
chr12_-_47679176
|
1.888
|
NM_015086
|
DDN
|
dendrin
|
|
chr9_+_966963
|
1.877
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr5_+_5193442
|
1.871
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr14_+_73775927
|
1.852
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr19_+_52215982
|
1.823
|
NM_002517
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr3_+_214668
|
1.821
|
|
|
|
|
chr18_+_48120539
|
1.792
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr9_-_84867604
|
1.785
|
|
RASEF
|
RAS and EF-hand domain containing
|
|
chr13_-_83354474
|
1.784
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr15_+_82113813
|
1.767
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr16_-_47872973
|
1.759
|
|
CBLN1
|
cerebellin 1 precursor
|
|
chr3_-_13436725
|
1.748
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr5_+_170779664
|
1.742
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr11_-_407324
|
1.742
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr9_-_99740243
|
1.740
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr17_-_71747867
|
1.735
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr19_+_2240996
|
1.734
|
|
|
|
|
chr13_+_40929525
|
1.721
|
NM_014059
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr13_-_83354425
|
1.721
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr4_+_2390469
|
1.705
|
NM_001193282
|
LOC402160
|
hypothetical protein LOC402160
|
|
chr3_+_127743868
|
1.701
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr5_+_88221013
|
1.701
|
|
RPS6
|
ribosomal protein S6
|
|
chr12_-_42231990
|
1.695
|
NM_025003
|
ADAMTS20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20
|
|
chr1_+_1557419
|
1.681
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr15_+_46271158
|
1.677
|
NM_001145668
|
CTXN2
SLC12A1
|
cortexin 2
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr2_-_233060774
|
1.658
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr15_+_62230962
|
1.640
|
NM_024798
|
SNX22
|
sorting nexin 22
|
|
chr6_-_166641860
|
1.631
|
NM_175922
|
PRR18
|
proline rich 18
|
|
chr9_+_134843918
|
1.628
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr17_-_7106497
|
1.622
|
|
CLDN7
|
claudin 7
|
|
chr13_+_40929690
|
1.618
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr13_+_40929679
|
1.606
|
|
C13orf15
|
chromosome 13 open reading frame 15
|
|
chr15_-_81113676
|
1.604
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr19_-_7845325
|
1.593
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr1_+_226937732
|
1.589
|
|
RHOU
|
ras homolog gene family, member U
|
|
chr14_+_61653827
|
1.589
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr1_+_95355415
|
1.575
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr3_-_193928038
|
1.564
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr5_-_115938305
|
1.551
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr12_+_93066370
|
1.526
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr9_-_10602722
|
1.517
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr3_-_13436802
|
1.516
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr15_+_31390454
|
1.514
|
NM_001036
|
RYR3
|
ryanodine receptor 3
|
|
chr17_+_19853200
|
1.513
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr17_-_8809656
|
1.495
|
NM_001142633
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5
|
|
chr10_+_106390848
|
1.488
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr10_+_72826656
|
1.466
|
NM_001171930
NM_001171931
NM_001171932
NM_022124
NM_052836
|
CDH23
|
cadherin-related 23
|
|
chr16_-_47872880
|
1.452
|
|
|
|
|
chr2_-_27195276
|
1.423
|
NM_001166239
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr11_+_45900731
|
1.389
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr14_+_41146513
|
1.381
|
NM_152447
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5
|
|
chr13_+_112704077
|
1.375
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr1_+_12149637
|
1.357
|
NM_001066
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr19_+_3536541
|
1.352
|
NM_133261
|
GIPC3
|
GIPC PDZ domain containing family, member 3
|
|
chr16_-_47873183
|
1.325
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chrX_-_142550535
|
1.322
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr15_-_80123935
|
1.317
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr12_-_47679459
|
1.315
|
|
DDN
|
dendrin
|
|
chr1_+_82038669
|
1.315
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr4_+_91267706
|
1.310
|
NM_001145065
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr10_-_98470164
|
1.305
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr4_-_21559471
|
1.303
|
NM_147182
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr1_+_175407153
|
1.288
|
NM_021165
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr2_-_133144094
|
1.285
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr16_+_3002457
|
1.284
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr8_+_26427378
|
1.279
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr11_-_45264193
|
1.277
|
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr1_-_194844061
|
1.263
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr17_-_34084682
|
1.261
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr9_-_135029096
|
1.260
|
NM_021996
|
GBGT1
RALGDS
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1
ral guanine nucleotide dissociation stimulator
|
|
chr16_-_716367
|
1.259
|
NM_001031737
|
CCDC78
|
coiled-coil domain containing 78
|
|
chr19_-_14177980
|
1.259
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr2_-_6924092
|
1.247
|
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr9_+_89302239
|
1.243
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr13_+_24152548
|
1.235
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr6_-_40663089
|
1.228
|
NM_020737
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr1_+_12149662
|
1.222
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr15_-_50869379
|
1.210
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chr4_+_177223978
|
1.203
|
NM_170710
NM_181265
|
WDR17
|
WD repeat domain 17
|
|
chr18_+_61569136
|
1.201
|
NM_004361
|
CDH7
|
cadherin 7, type 2
|
|
chr6_+_99389300
|
1.200
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr5_+_146594718
|
1.198
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr8_-_57521702
|
1.197
|
NM_001135690
|
PENK
|
proenkephalin
|
|
chr4_+_177224142
|
1.197
|
|
WDR17
|
WD repeat domain 17
|
|
chr5_+_88220846
|
1.193
|
|
|
|
|
chr22_+_28446288
|
1.191
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr4_-_41579325
|
1.185
|
|
|
|
|
chr17_+_5914364
|
1.183
|
NM_015253
|
WSCD1
|
WSC domain containing 1
|
|
chr2_+_112372671
|
1.181
|
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr4_-_175680366
|
1.181
|
NM_000860
NM_001145816
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr13_+_112704051
|
1.179
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr7_+_156496360
|
1.177
|
|
LOC645249
|
hypothetical LOC645249
|
|
chr21_-_31853160
|
1.170
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr12_+_79634820
|
1.162
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr13_+_112704028
|
1.160
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr11_-_33847507
|
1.160
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr12_-_31163309
|
1.142
|
|
|
|
|
chr15_-_75694674
|
1.140
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr16_+_65754788
|
1.137
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr19_-_62912347
|
1.137
|
NM_001085384
|
ZNF154
|
zinc finger protein 154
|
|
chr5_-_132976121
|
1.134
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr17_-_43862598
|
1.131
|
|
SKAP1
|
src kinase associated phosphoprotein 1
|
|
chr2_-_100088224
|
1.126
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr9_-_23811477
|
1.126
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr18_+_48121067
|
1.126
|
|
DCC
|
deleted in colorectal carcinoma
|
|
chr6_+_133604181
|
1.124
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr5_-_139992621
|
1.124
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr11_-_16992524
|
1.121
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr14_+_71468902
|
1.120
|
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr8_+_120497732
|
1.114
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr19_-_54618410
|
1.105
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chr1_+_154605626
|
1.102
|
NM_020407
|
RHBG
|
Rh family, B glycoprotein (gene/pseudogene)
|
|
chr19_-_18867868
|
1.098
|
NM_021267
NM_198207
NM_001492
|
LASS1
GDF1
|
LAG1 homolog, ceramide synthase 1
growth differentiation factor 1
|
|
chr11_-_118799452
|
1.094
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr17_-_76064842
|
1.090
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr1_-_86394695
|
1.088
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chr8_+_21937533
|
1.079
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr12_-_73889777
|
1.077
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr6_+_19946591
|
1.075
|
|
|
|
|
chr10_+_106391032
|
1.073
|
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr5_-_95793618
|
1.062
|
NM_001177875
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr13_-_52320774
|
1.059
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr6_+_133604203
|
1.054
|
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr2_-_74580173
|
1.053
|
|
LBX2
|
ladybird homeobox 2
|
|
chr15_-_86600658
|
1.051
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr17_-_7106987
|
1.051
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr20_+_13150417
|
1.051
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr20_+_20296744
|
1.050
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr16_+_1323627
|
1.042
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr5_+_31229549
|
1.038
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr3_-_112796855
|
1.036
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr4_-_11039600
|
1.017
|
NM_005114
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1
|
|
chr2_-_166940710
|
1.016
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr19_-_2259155
|
1.012
|
NM_001101391
|
LINGO3
|
leucine rich repeat and Ig domain containing 3
|
|
chr2_-_241408296
|
1.011
|
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr1_-_194844299
|
1.011
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr6_-_89884324
|
1.006
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr3_-_129777618
|
1.002
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr11_+_1367665
|
0.973
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr4_+_156900053
|
0.964
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr19_-_18198234
|
0.959
|
NM_001098818
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|