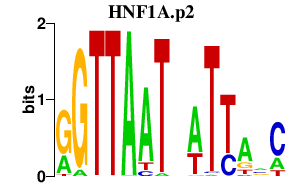
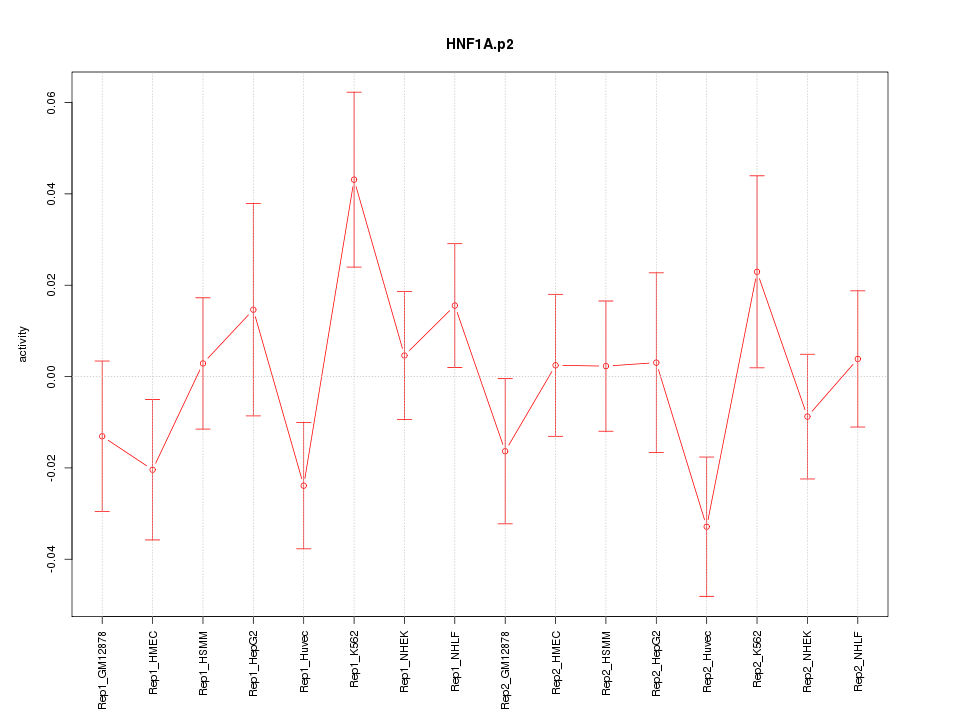
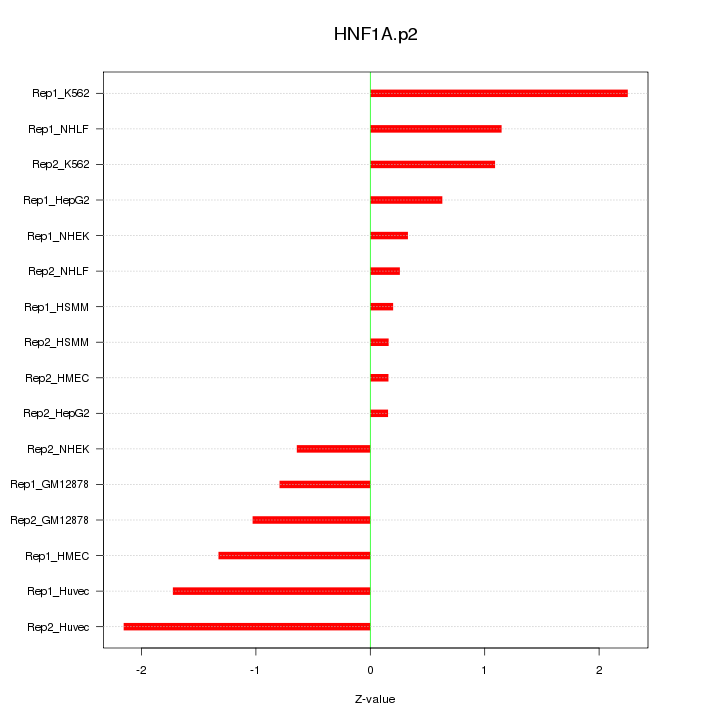
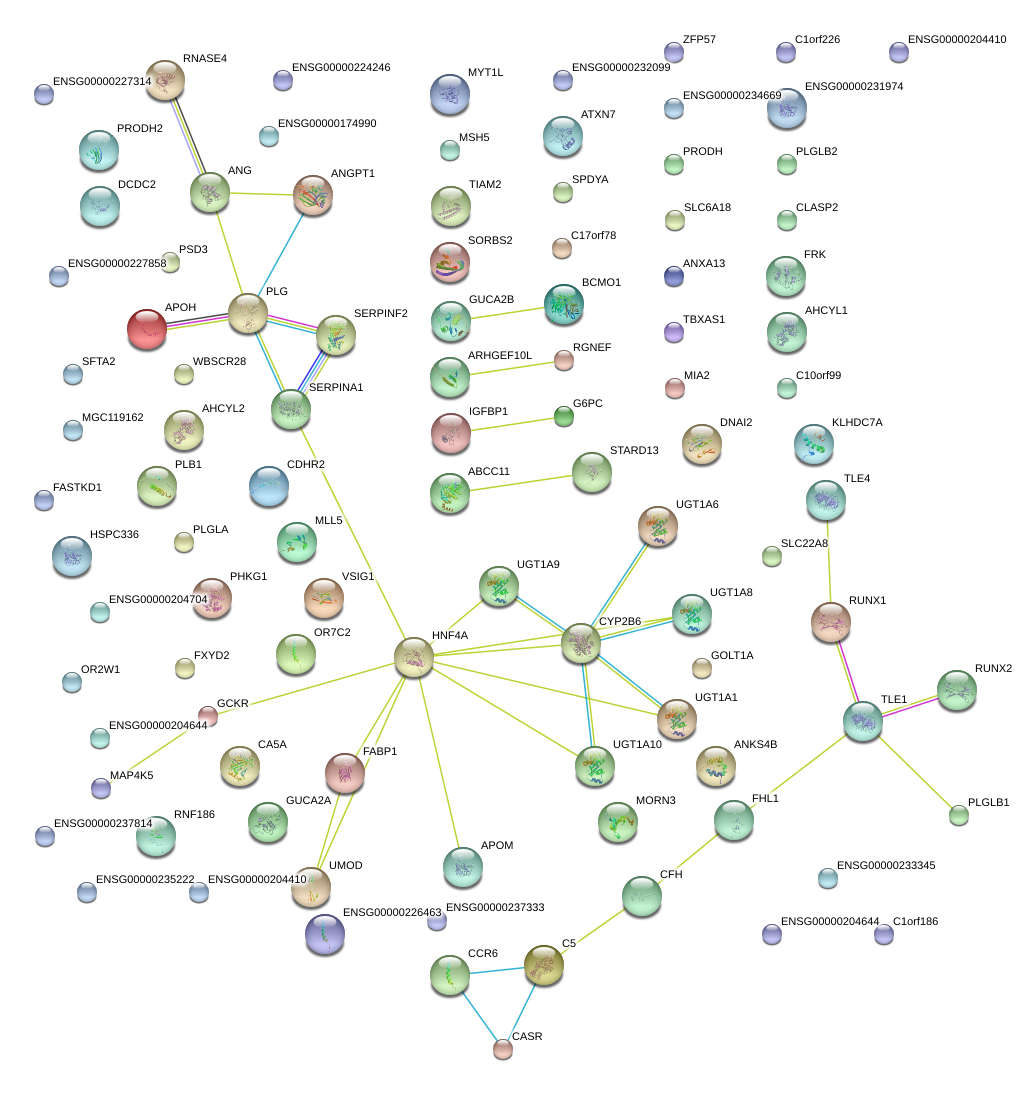
|
chr6_-_24491498
|
2.161
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr11_-_117200618
|
2.026
|
NM_001127489
NM_001680
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr5_+_1278469
|
1.654
|
NM_182632
|
SLC6A18
|
solute carrier family 6, member 18
|
|
chr19_-_40995564
|
1.326
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr1_-_20014180
|
1.307
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chr14_+_20226762
|
1.293
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr17_-_61655915
|
1.221
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr1_+_18680010
|
1.167
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr1_-_204455269
|
1.112
|
NM_001007544
|
C1orf186
|
chromosome 1 open reading frame 186
|
|
chr5_+_73145098
|
1.104
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr2_+_234333653
|
1.097
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr6_-_29752909
|
1.095
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr5_+_175908948
|
1.091
|
NM_017675
|
CDHR2
|
cadherin-related family member 2
|
|
chr19_-_40995606
|
1.073
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr16_-_86527608
|
1.063
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr10_+_85923473
|
1.012
|
NM_207373
|
C10orf99
|
chromosome 10 open reading frame 99
|
|
chrX_+_135079120
|
1.009
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_+_31815703
|
1.007
|
NM_002441
NM_025259
NM_172165
NM_172166
|
MSH5-C6ORF26
MSH5
|
MSH5-C6orf26 read-through transcript
mutS homolog 5 (E. coli)
|
|
chr6_-_31007905
|
0.970
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr1_+_17779528
|
0.962
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr2_-_87102453
|
0.951
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr2_+_27573206
|
0.934
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr7_+_45894480
|
0.931
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr8_-_124818754
|
0.891
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr8_-_18585721
|
0.889
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_+_42391678
|
0.882
|
NM_007102
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin)
|
|
chr13_-_32658215
|
0.859
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_-_186970366
|
0.858
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr20_+_42417752
|
0.858
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr7_+_128802719
|
0.845
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_42402973
|
0.836
|
NM_033553
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin)
|
|
chr6_-_29120930
|
0.819
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chr7_+_139175420
|
0.813
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr16_+_79829794
|
0.791
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr6_-_116488458
|
0.774
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr7_-_104354325
|
0.769
|
|
LOC723809
|
hypothetical LOC723809
|
|
chr2_-_88208650
|
0.750
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr6_+_161043214
|
0.714
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr16_-_54466709
|
0.711
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr19_+_46201665
|
0.709
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr19_+_14913298
|
0.685
|
NM_012377
|
OR7C2
|
olfactory receptor, family 7, subfamily C, member 2
|
|
chr2_+_87828718
|
0.683
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr6_+_31081707
|
0.669
|
NM_001198815
|
PBMUCL1
|
panbronchiolitis related mucin-like 1
|
|
chr7_+_72913424
|
0.656
|
NM_182504
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr13_+_50813168
|
0.649
|
NM_001101320
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3
|
|
chr20_+_42417502
|
0.642
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_-_1925880
|
0.627
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr2_+_28572441
|
0.623
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr2_+_28892366
|
0.610
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr22_+_21027538
|
0.602
|
|
|
|
|
chr2_+_234191029
|
0.595
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr17_+_38306339
|
0.589
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr7_+_104539548
|
0.589
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr6_+_31731647
|
0.586
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chrX_+_107174855
|
0.585
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr11_-_62539857
|
0.584
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr16_+_21152516
|
0.579
|
NM_145865
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr16_-_20271530
|
0.570
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr17_+_69781980
|
0.557
|
NM_001172810
NM_023036
|
DNAI2
|
dynein, axonemal, intermediate chain 2
|
|
chr21_-_35153664
|
0.543
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_+_31731667
|
0.541
|
|
APOM
|
apolipoprotein M
|
|
chr9_-_122852356
|
0.516
|
NM_001735
|
C5
|
complement component 5
|
|
chr10_+_99334411
|
0.508
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr9_+_81376666
|
0.505
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr3_-_33675484
|
0.501
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr14_-_93924663
|
0.500
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr3_+_63928459
|
0.499
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr16_-_46826588
|
0.493
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr20_+_42463419
|
0.479
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr8_-_108579270
|
0.468
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr6_+_155453114
|
0.460
|
NM_012454
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr6_+_167456230
|
0.453
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr1_-_202449659
|
0.451
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr1_+_160618143
|
0.449
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr2_-_170138558
|
0.442
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr17_+_1592879
|
0.427
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr17_+_32807040
|
0.398
|
NM_173625
|
C17orf78
|
chromosome 17 open reading frame 78
|
|
chr14_+_38772870
|
0.391
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr20_+_42463387
|
0.389
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr15_-_49184764
|
0.388
|
NM_207381
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr19_+_11211282
|
0.387
|
NM_018687
|
LOC55908
|
hepatocellular carcinoma-associated gene TD26
|
|
chr2_-_1905508
|
0.377
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr10_-_101680645
|
0.366
|
|
DNMBP
|
dynamin binding protein
|
|
chr10_-_32707549
|
0.363
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr6_+_29249289
|
0.352
|
NM_030905
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2
|
|
chr14_-_93924873
|
0.350
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr15_+_60726801
|
0.350
|
NM_015059
|
TLN2
|
talin 2
|
|
chr4_-_72868598
|
0.349
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr3_+_188397967
|
0.345
|
NM_153708
|
RTP1
|
receptor (chemosensory) transporter protein 1
|
|
chr17_-_10393664
|
0.344
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr4_+_56731117
|
0.341
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chr2_+_151922350
|
0.339
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr7_-_27133163
|
0.333
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr3_+_187866464
|
0.332
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr20_+_42463350
|
0.331
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr7_-_27186400
|
0.312
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr2_+_101981888
|
0.308
|
NM_173343
|
IL1R2
|
interleukin 1 receptor, type II
|
|
chr17_+_71154240
|
0.301
|
NM_001162997
|
LOC100130933
|
hypothetical protein LOC100130933
|
|
chr6_+_158990982
|
0.300
|
NM_001009991
|
SYTL3
|
synaptotagmin-like 3
|
|
chr7_-_87694218
|
0.300
|
NM_198901
|
SRI
|
sorcin
|
|
chr1_+_109057078
|
0.295
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr3_-_168674502
|
0.294
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr7_-_107667728
|
0.288
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr17_+_1592893
|
0.287
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr3_-_25654659
|
0.287
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr10_+_69539255
|
0.284
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_-_171070314
|
0.281
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr6_+_121798443
|
0.280
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr4_-_102487649
|
0.278
|
NM_000944
NM_001130691
NM_001130692
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr11_+_64114857
|
0.274
|
NM_144585
NM_153378
|
SLC22A12
|
solute carrier family 22 (organic anion/urate transporter), member 12
|
|
chrX_+_70351773
|
0.272
|
NM_001097642
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr6_+_155579788
|
0.272
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr1_-_246071820
|
0.270
|
NM_001001959
|
OR11L1
|
olfactory receptor, family 11, subfamily L, member 1
|
|
chrX_+_70351812
|
0.265
|
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr2_-_99284070
|
0.261
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr5_+_147775652
|
0.261
|
|
FBXO38
|
F-box protein 38
|
|
chr1_-_157950898
|
0.260
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr12_-_122400937
|
0.258
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr6_-_26038817
|
0.252
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr5_+_156540425
|
0.246
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr19_-_38052481
|
0.244
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr7_-_107930170
|
0.244
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr20_+_42463279
|
0.244
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_+_234266250
|
0.240
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr6_+_64289585
|
0.239
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr17_+_76549658
|
0.239
|
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr14_+_64240996
|
0.231
|
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr1_-_114231691
|
0.230
|
NM_001010922
|
BCL2L15
|
BCL2-like 15
|
|
chr7_-_50600536
|
0.228
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr5_+_67622242
|
0.223
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr21_-_42219849
|
0.222
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr7_-_107230867
|
0.222
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr6_-_29563657
|
0.221
|
NM_052967
|
MAS1L
|
MAS1 oncogene-like
|
|
chr17_-_24253821
|
0.218
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr5_-_138746840
|
0.217
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr20_+_61841553
|
0.215
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr14_+_19473606
|
0.214
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr10_+_99334069
|
0.214
|
NM_001134670
NM_138413
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr1_-_67039518
|
0.211
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr2_+_62787534
|
0.211
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr2_-_63518589
|
0.205
|
NM_001042692
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr14_-_93859394
|
0.202
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr12_-_54965996
|
0.202
|
|
CS
|
citrate synthase
|
|
chr15_-_96875133
|
0.198
|
NM_182562
|
FAM169B
|
family with sequence similarity 169, member B
|
|
chr12_+_91620749
|
0.197
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr2_+_101874809
|
0.196
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr17_-_53335748
|
0.195
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr4_-_69852090
|
0.194
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr11_-_101292462
|
0.194
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr4_+_187424111
|
0.192
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr12_+_10222823
|
0.191
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr1_+_184532027
|
0.191
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr1_+_55044323
|
0.190
|
NM_001110533
NM_152607
|
C1orf177
|
chromosome 1 open reading frame 177
|
|
chr19_-_40996040
|
0.188
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr1_-_228916559
|
0.187
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr8_-_17797127
|
0.186
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr10_+_18669619
|
0.185
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr14_+_95915774
|
0.185
|
NM_016472
|
C14orf129
|
chromosome 14 open reading frame 129
|
|
chr10_-_69736600
|
0.184
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr10_+_35466715
|
0.181
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr6_-_108252200
|
0.178
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr4_-_74704964
|
0.178
|
NM_177532
NM_201431
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr2_+_234302511
|
0.175
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr17_+_1593040
|
0.175
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr12_-_122322117
|
0.174
|
NM_004642
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1
|
|
chrX_-_15593074
|
0.171
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr3_-_158704056
|
0.169
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr10_-_85921703
|
0.165
|
|
LOC642934
|
hypothetical LOC642934
|
|
chr16_+_88224276
|
0.164
|
|
DPEP1
|
dipeptidase 1 (renal)
|
|
chr6_-_33862690
|
0.163
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr1_+_93830169
|
0.162
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr22_-_30981317
|
0.162
|
NM_014227
|
SLC5A4
|
solute carrier family 5 (low affinity glucose cotransporter), member 4
|
|
chr20_+_35406497
|
0.160
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr11_-_67207112
|
0.160
|
|
RPL37
|
ribosomal protein L37
|
|
chr19_-_11500912
|
0.159
|
NM_001142464
NM_001142465
NM_016581
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr2_+_182558795
|
0.158
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr17_-_19290745
|
0.157
|
|
|
|
|
chr5_-_39430180
|
0.156
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr4_-_189263395
|
0.153
|
NM_173553
|
TRIML2
|
tripartite motif family-like 2
|
|
chr1_+_93830106
|
0.153
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr21_+_38550739
|
0.152
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr7_-_36992221
|
0.152
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr8_+_98772537
|
0.151
|
|
MTDH
|
metadherin
|
|
chr11_-_8695974
|
0.151
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr15_+_83724743
|
0.151
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr11_-_123130435
|
0.146
|
NM_001005188
|
OR6X1
|
olfactory receptor, family 6, subfamily X, member 1
|
|
chr18_+_21967813
|
0.144
|
NM_001025096
NM_001025097
NM_144662
|
PSMA8
|
proteasome (prosome, macropain) subunit, alpha type, 8
|
|
chr13_-_99983945
|
0.144
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr10_+_101532450
|
0.141
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr16_-_67943157
|
0.139
|
NM_144676
|
TMED6
|
transmembrane emp24 protein transport domain containing 6
|
|
chr4_+_154345039
|
0.139
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr13_-_98757634
|
0.138
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr1_-_195818927
|
0.137
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr1_-_9052253
|
0.136
|
NM_001135585
NM_003039
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr7_-_44547382
|
0.133
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr4_+_187227302
|
0.132
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr15_+_22772545
|
0.132
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr1_-_114231519
|
0.132
|
|
BCL2L15
|
BCL2-like 15
|
|
chr3_-_57209319
|
0.131
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chrX_+_107569671
|
0.129
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|