|
chr6_+_106653429
|
3.035
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_50789215
|
2.386
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr1_-_116184846
|
2.032
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr10_-_45131061
|
1.662
|
NM_001004297
|
OR13A1
|
olfactory receptor, family 13, subfamily A, member 1
|
|
chr2_+_202379442
|
1.628
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr12_+_54011750
|
1.619
|
NM_054104
|
OR6C3
|
olfactory receptor, family 6, subfamily C, member 3
|
|
chr12_-_8584658
|
1.579
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr5_-_88004891
|
1.572
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr4_-_122357104
|
1.557
|
NM_001128843
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr20_+_1132097
|
1.555
|
NM_001009612
|
C20orf202
|
chromosome 20 open reading frame 202
|
|
chr10_+_111755715
|
1.525
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr1_-_150564302
|
1.452
|
NM_002016
|
FLG
|
filaggrin
|
|
chr12_+_99274987
|
1.452
|
NM_001145288
NM_139319
|
SLC17A8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8
|
|
chr5_-_135259403
|
1.426
|
NM_000590
|
IL9
|
interleukin 9
|
|
chr11_-_40271086
|
1.414
|
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr5_+_140690387
|
1.389
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr6_+_127940011
|
1.371
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr3_-_150533948
|
1.292
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr17_-_36074812
|
1.277
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr2_-_200038055
|
1.265
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr1_+_24518398
|
1.260
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr10_-_51678318
|
1.251
|
NM_001143974
NM_019893
|
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2
|
|
chrX_-_15593074
|
1.210
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr1_+_169484256
|
1.207
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr15_+_60838053
|
1.197
|
|
TLN2
|
talin 2
|
|
chr12_-_9976173
|
1.193
|
NM_001130711
|
CLEC2A
|
C-type lectin domain family 2, member A
|
|
chr21_-_38792173
|
1.164
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_+_90336489
|
1.145
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr14_+_85066265
|
1.121
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr6_+_132901119
|
1.116
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr4_+_154083584
|
1.095
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr2_-_134042500
|
1.091
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr1_-_67039518
|
1.081
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr4_+_71283383
|
1.073
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr4_+_15080586
|
1.071
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr17_+_7283412
|
1.063
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr4_-_174774368
|
1.059
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr5_-_16561936
|
1.051
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr6_+_129245978
|
1.030
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr5_+_27508139
|
1.021
|
|
LOC643401
|
hypothetical protein LOC643401
|
|
chr11_+_118259684
|
1.020
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr3_-_125192888
|
1.017
|
NM_017578
|
ROPN1
|
rhophilin associated tail protein 1
|
|
chr8_-_30826074
|
1.007
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr1_-_203657803
|
1.004
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr8_-_13416554
|
0.984
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_+_99465777
|
0.981
|
NM_001005479
|
OR5H6
|
olfactory receptor, family 5, subfamily H, member 6
|
|
chr10_-_115413630
|
0.967
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr15_+_41672543
|
0.946
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr9_-_72673753
|
0.936
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr2_+_168383427
|
0.936
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr7_+_123353144
|
0.935
|
NM_001174045
NM_001174046
|
SPAM1
|
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding)
|
|
chr11_-_124311512
|
0.930
|
NM_152722
|
HEPACAM
|
hepatic and glial cell adhesion molecule
|
|
chr14_-_20694023
|
0.917
|
NM_001004731
|
OR5AU1
|
olfactory receptor, family 5, subfamily AU, member 1
|
|
chr7_-_83116414
|
0.916
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr14_+_75907442
|
0.913
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr9_-_92444886
|
0.910
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr6_+_132000134
|
0.910
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr3_-_159306214
|
0.903
|
|
|
|
|
chr6_+_123142559
|
0.902
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr10_+_63092724
|
0.897
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr4_+_130236731
|
0.891
|
NM_001099783
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr1_-_109201228
|
0.889
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr9_-_14900992
|
0.887
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr10_-_100985548
|
0.884
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr21_+_38550739
|
0.884
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr4_-_120462705
|
0.883
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr1_+_239882202
|
0.882
|
NM_144625
|
WDR64
|
WD repeat domain 64
|
|
chr2_+_66519949
|
0.878
|
|
MEIS1
|
Meis homeobox 1
|
|
chr13_-_33148899
|
0.867
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_-_155936904
|
0.854
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr17_-_23244499
|
0.852
|
NM_001076680
|
C17orf108
|
chromosome 17 open reading frame 108
|
|
chr8_+_32698892
|
0.849
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr2_-_179623030
|
0.846
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr3_-_27739201
|
0.842
|
|
EOMES
|
eomesodermin
|
|
chr11_+_22316242
|
0.838
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr17_-_51155073
|
0.827
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr18_+_57308754
|
0.827
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr1_+_148496792
|
0.821
|
NM_012113
|
CA14
|
carbonic anhydrase XIV
|
|
chr4_+_88187215
|
0.809
|
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr1_-_146422000
|
0.809
|
NM_178230
|
PPIAL4A
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
|
|
chr1_+_47261826
|
0.804
|
NM_178033
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1
|
|
chr5_+_132176921
|
0.804
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr11_+_101488380
|
0.792
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr13_+_22653059
|
0.792
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr11_-_5302157
|
0.791
|
NM_033180
|
OR51B2
|
olfactory receptor, family 51, subfamily B, member 2
|
|
chr21_-_26867138
|
0.791
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr3_-_162572441
|
0.782
|
NM_001040100
|
C3orf57
|
chromosome 3 open reading frame 57
|
|
chr1_-_79244948
|
0.780
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr10_-_50269912
|
0.780
|
NM_001080520
|
DRGX
|
dorsal root ganglia homeobox
|
|
chr19_-_47623340
|
0.779
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr6_+_123142344
|
0.779
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr5_+_152850276
|
0.778
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr8_-_17811750
|
0.776
|
|
FGL1
|
fibrinogen-like 1
|
|
chr4_-_101658094
|
0.765
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr4_+_71492581
|
0.764
|
NM_016519
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr6_-_136829705
|
0.763
|
NM_001198615
|
MAP7
|
microtubule-associated protein 7
|
|
chr20_-_1113116
|
0.762
|
NM_018354
|
C20orf46
|
chromosome 20 open reading frame 46
|
|
chr11_+_89507465
|
0.761
|
NM_005467
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2
|
|
chr6_-_89984214
|
0.754
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr3_-_152529999
|
0.751
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr3_+_99592199
|
0.744
|
NM_001005516
|
OR5K3
|
olfactory receptor, family 5, subfamily K, member 3
|
|
chr6_-_11887265
|
0.741
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr6_-_69401639
|
0.737
|
|
|
|
|
chr1_-_155281785
|
0.736
|
NM_014784
NM_198236
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11
|
|
chr21_-_41479035
|
0.735
|
NM_182832
|
PLAC4
|
placenta-specific 4
|
|
chr14_-_21074875
|
0.732
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr11_-_5247948
|
0.731
|
NM_005330
|
HBE1
|
hemoglobin, epsilon 1
|
|
chr6_-_47117951
|
0.729
|
NM_025048
NM_153840
|
GPR110
|
G protein-coupled receptor 110
|
|
chr14_+_61532293
|
0.729
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr1_+_169326641
|
0.728
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr5_+_76284435
|
0.724
|
NM_001882
|
CRHBP
|
corticotropin releasing hormone binding protein
|
|
chr14_-_68424653
|
0.720
|
|
ACTN1
|
actinin, alpha 1
|
|
chr4_-_123761615
|
0.718
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr1_+_171871283
|
0.717
|
NM_001195190
|
LOC730159
|
hypothetical protein LOC730159
|
|
chrX_+_135558001
|
0.715
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr17_-_64462976
|
0.709
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr21_-_42060334
|
0.705
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr1_+_147819626
|
0.704
|
NM_178230
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr1_+_156991133
|
0.704
|
NM_001005184
|
OR6K6
|
olfactory receptor, family 6, subfamily K, member 6
|
|
chr2_-_172675873
|
0.703
|
|
DLX2
|
distal-less homeobox 2
|
|
chr14_-_98807307
|
0.700
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr11_+_129484540
|
0.700
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr15_+_22652790
|
0.698
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr12_+_52696908
|
0.696
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr8_+_100025806
|
0.694
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr1_+_24518501
|
0.691
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr18_-_51219880
|
0.687
|
|
TCF4
|
transcription factor 4
|
|
chr5_-_59100194
|
0.687
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_100645032
|
0.685
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr17_-_36403910
|
0.684
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr15_+_41772375
|
0.684
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chrX_+_128700605
|
0.680
|
NM_003399
|
XPNPEP2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound
|
|
chr8_+_120289735
|
0.677
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr14_+_98247702
|
0.676
|
NM_182560
|
C14orf177
|
chromosome 14 open reading frame 177
|
|
chr6_+_116956868
|
0.671
|
NM_153036
|
FAM26D
|
family with sequence similarity 26, member D
|
|
chr12_-_45506001
|
0.670
|
NM_001143824
NM_018018
|
SLC38A4
|
solute carrier family 38, member 4
|
|
chr3_-_18455207
|
0.670
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr1_-_142559339
|
0.670
|
NM_001123068
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G
|
|
chr11_+_5597749
|
0.666
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr2_-_162808123
|
0.653
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr8_-_17797127
|
0.651
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chrX_-_31194867
|
0.648
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr22_-_35733784
|
0.648
|
NM_001163857
NM_178552
|
C22orf33
|
chromosome 22 open reading frame 33
|
|
chr14_-_23623657
|
0.646
|
NM_006177
|
NRL
|
neural retina leucine zipper
|
|
chr13_+_27392136
|
0.644
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr9_+_122890394
|
0.643
|
NM_007018
|
CEP110
|
centrosomal protein 110kDa
|
|
chr4_-_5071996
|
0.642
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr11_-_123695302
|
0.641
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr3_+_29297806
|
0.640
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr12_+_4700012
|
0.640
|
NM_017417
|
GALNT8
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 8 (GalNAc-T8)
|
|
chr2_-_200028977
|
0.637
|
|
SATB2
|
SATB homeobox 2
|
|
chr1_-_143075560
|
0.635
|
NM_178230
NM_001143883
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4B
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4B
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr2_-_142605739
|
0.633
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr8_+_7340777
|
0.633
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr20_+_5839973
|
0.633
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr12_-_28016182
|
0.633
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr10_-_134449467
|
0.632
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr18_+_54013575
|
0.631
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr4_-_122304925
|
0.628
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr13_+_23042722
|
0.625
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr4_+_74520796
|
0.625
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr17_+_57811645
|
0.621
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr5_-_16791346
|
0.615
|
|
MYO10
|
myosin X
|
|
chr11_+_56187713
|
0.613
|
NM_001004730
|
OR5AR1
|
olfactory receptor, family 5, subfamily AR, member 1
|
|
chr7_-_27133163
|
0.607
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr10_-_90601655
|
0.607
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr3_+_148609841
|
0.607
|
NM_003412
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr4_+_159662349
|
0.605
|
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr12_+_7833292
|
0.605
|
|
NANOG
|
Nanog homeobox
|
|
chr11_+_5367182
|
0.605
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr5_+_180269169
|
0.604
|
NM_001159710
|
BTNL8
|
butyrophilin-like 8
|
|
chr5_-_135318420
|
0.603
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr2_-_213723298
|
0.601
|
NM_016260
|
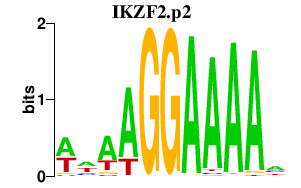
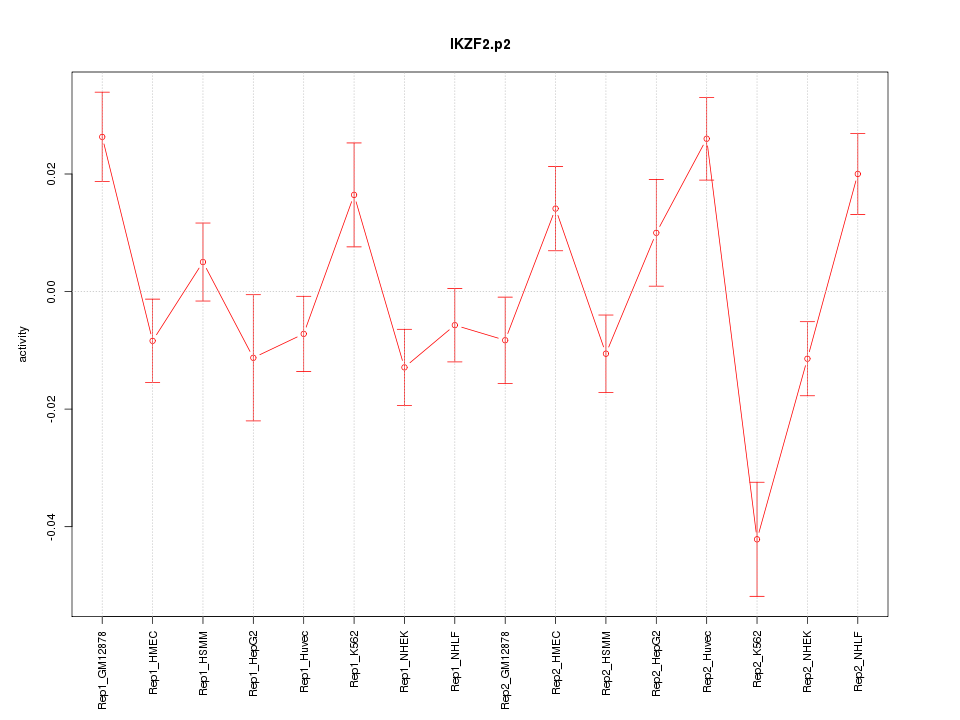
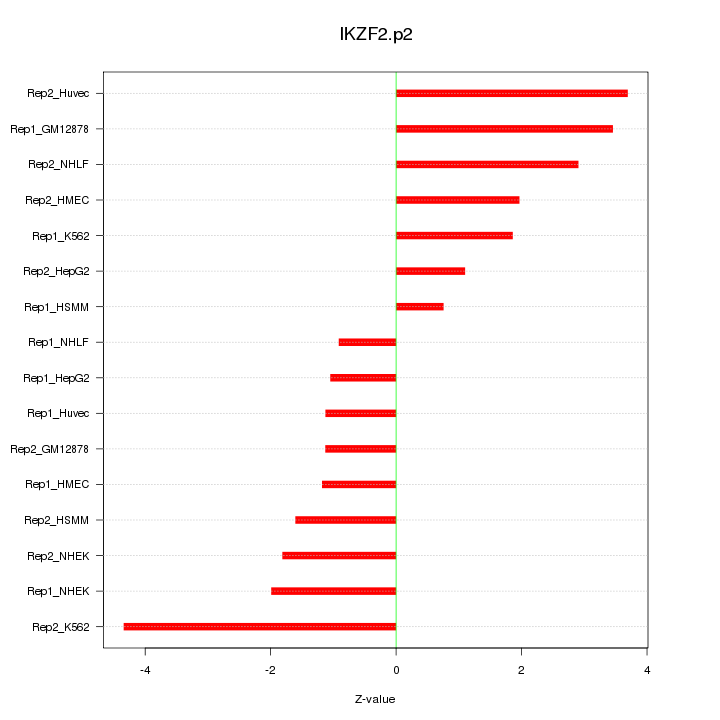
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_115415061
|
0.601
|
NM_001195581
|
LOC644100
|
hypothetical LOC644100
|
|
chr4_-_77547481
|
0.597
|
NM_001042784
|
CCDC158
|
coiled-coil domain containing 158
|
|
chr1_+_61642287
|
0.596
|
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_171838802
|
0.596
|
NM_178527
|
SLC9A11
|
solute carrier family 9, member 11
|
|
chr8_-_82606104
|
0.594
|
NM_001105281
|
FABP12
|
fatty acid binding protein 12
|
|
chr3_+_134985566
|
0.594
|
NM_021203
|
SRPRB
|
signal recognition particle receptor, B subunit
|
|
chr6_-_101018271
|
0.593
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr14_+_85066207
|
0.593
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr11_+_18374388
|
0.588
|
NM_001165414
|
LDHA
|
lactate dehydrogenase A
|
|
chr9_-_21177529
|
0.584
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr1_-_151333624
|
0.584
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr11_-_57171554
|
0.581
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr5_-_115938305
|
0.580
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr4_-_100575313
|
0.580
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr1_-_36338436
|
0.579
|
NM_005202
|
COL8A2
|
collagen, type VIII, alpha 2
|
|
chrX_-_74292800
|
0.579
|
NM_004299
|
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7
|
|
chr8_-_82522175
|
0.577
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr11_+_76423082
|
0.575
|
NM_138706
|
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase)
|
|
chr12_+_54231249
|
0.574
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chr2_-_189752575
|
0.573
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_+_50119261
|
0.572
|
|
RBM5
|
RNA binding motif protein 5
|
|
chr9_-_15240113
|
0.569
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr10_+_104580420
|
0.568
|
|
|
|
|
chr8_+_102574012
|
0.568
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr1_+_146910634
|
0.565
|
NM_001144032
|
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr14_-_31026901
|
0.562
|
NM_001197184
|
GPR33
|
G protein-coupled receptor 33 (gene/pseudogene)
|
|
chr9_-_21358005
|
0.562
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|