|
chr10_-_103525646
|
6.492
|
NM_006119
NM_033163
NM_033164
NM_033165
|
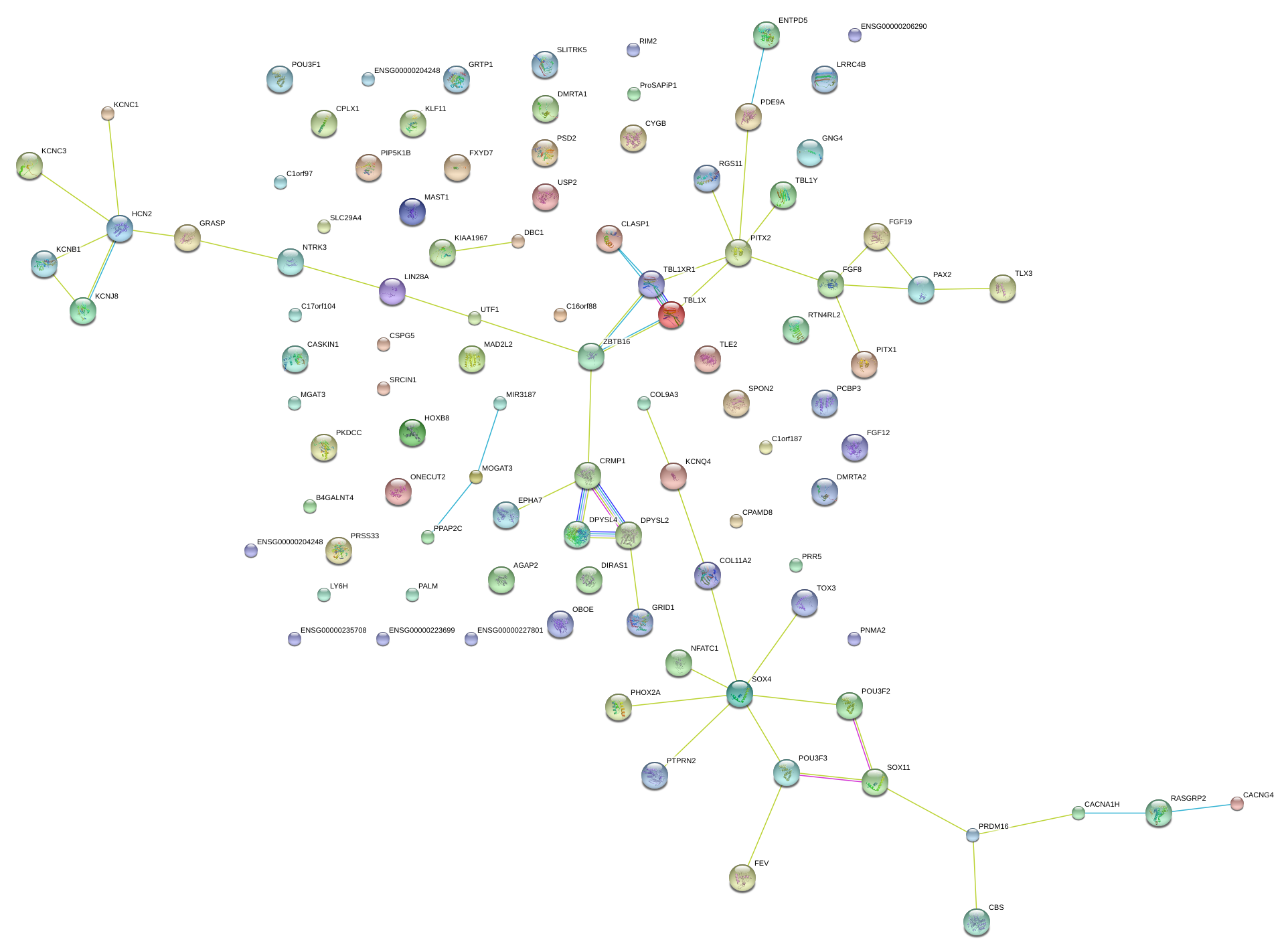
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr4_-_809879
|
6.355
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr19_+_659766
|
5.566
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr19_-_2672337
|
4.833
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr6_-_94185780
|
4.553
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr11_-_118739838
|
4.478
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr19_-_55524445
|
4.137
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr5_-_134397740
|
4.096
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr7_-_158073121
|
4.069
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr4_-_5940893
|
3.976
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr16_-_2186465
|
3.938
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr1_-_11674264
|
3.904
|
NM_001127325
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chr6_-_33268177
|
3.869
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr13_-_113066202
|
3.757
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr11_+_56984702
|
3.563
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr4_-_5945596
|
3.562
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr19_-_242335
|
3.543
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr7_+_5289079
|
3.421
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr11_-_118740069
|
3.350
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr15_-_86600658
|
3.222
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr17_-_44047299
|
3.221
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr1_-_233879617
|
3.198
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr8_-_144313100
|
3.192
|
NM_001135655
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr19_+_12810258
|
3.175
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr21_+_45888001
|
3.094
|
|
PCBP3
|
poly(rC) binding protein 3
|
|
chr11_+_359723
|
3.087
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr4_-_5941043
|
3.063
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr12_+_50686990
|
3.021
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr13_+_87122778
|
3.004
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr6_+_99389300
|
2.995
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr10_+_133850325
|
2.980
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr12_-_21818881
|
2.975
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr6_+_99389318
|
2.974
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr2_+_5750229
|
2.973
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr22_+_38183480
|
2.966
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr18_+_53253775
|
2.940
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr13_-_113066366
|
2.842
|
NM_024719
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr5_-_134397470
|
2.786
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr1_+_11674365
|
2.745
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr19_+_540849
|
2.718
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr3_-_47595190
|
2.687
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr19_-_16998449
|
2.648
|
NM_015692
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr8_+_104582151
|
2.617
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr10_+_134893766
|
2.615
|
NM_003577
|
UTF1
|
undifferentiated embryonic cell transcription factor 1
|
|
chr5_+_139155569
|
2.591
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr12_-_56418295
|
2.548
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr11_-_71632695
|
2.516
|
|
PHOX2A
|
paired-like homeobox 2a
|
|
chr11_+_17714070
|
2.510
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr1_+_26609819
|
2.503
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr20_-_3097070
|
2.455
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr4_-_111763655
|
2.441
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr1_+_2975590
|
2.434
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr19_-_55763091
|
2.405
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr4_-_1156362
|
2.393
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr16_-_265868
|
2.380
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr19_-_772966
|
2.369
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr10_+_102495327
|
2.358
|
NM_000278
NM_003987
NM_003988
NM_003989
NM_003990
|
PAX2
|
paired box 2
|
|
chr4_-_1156458
|
2.358
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr19_-_772913
|
2.358
|
NM_024888
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr8_-_144313053
|
2.356
|
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr2_+_10101081
|
2.327
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr10_-_88116181
|
2.326
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr4_-_1156533
|
2.321
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr19_+_40325993
|
2.319
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chr16_-_2776606
|
2.312
|
NM_152891
|
PRSS33
|
protease, serine, 33
|
|
chr4_-_1156378
|
2.289
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr1_+_41022066
|
2.241
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr21_+_42946719
|
2.231
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr21_-_43368963
|
2.222
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr9_+_70509925
|
2.221
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr17_+_40089270
|
2.214
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr22_+_43476710
|
2.151
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr11_-_69228286
|
2.145
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr13_+_99431417
|
2.136
|
|
|
|
|
chr19_-_53706644
|
2.123
|
|
|
|
|
chr4_-_5941167
|
2.117
|
NM_001313
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr2_-_219558517
|
2.114
|
NM_017521
|
FEV
|
FEV (ETS oncogene family)
|
|
chr20_-_47532585
|
2.105
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr1_+_26609893
|
2.097
|
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr16_+_1143241
|
2.090
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr9_-_121171400
|
2.090
|
NM_014618
|
DBC1
|
deleted in bladder cancer 1
|
|
chrX_+_9392980
|
2.086
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr19_-_2979901
|
2.083
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr17_+_62391216
|
2.083
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr16_-_51138306
|
2.079
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr1_-_29322993
|
2.072
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr20_+_60918836
|
2.065
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr11_-_71632864
|
2.056
|
NM_005169
|
PHOX2A
|
paired-like homeobox 2a
|
|
chr17_-_72045218
|
2.048
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr1_-_50661716
|
2.037
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr2_+_42128521
|
2.034
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr17_-_34014390
|
2.028
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr18_+_75256954
|
2.010
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_-_193609531
|
2.001
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr8_+_26427378
|
2.001
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_170668892
|
1.996
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr8_-_26427304
|
1.996
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr11_+_113435506
|
1.995
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr11_-_64268841
|
1.991
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr10_-_118021532
|
1.991
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr4_-_111763356
|
1.990
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr8_-_144313293
|
1.988
|
NM_001130478
|
LY6H
|
lymphocyte antigen 6 complex, locus H
|
|
chr5_-_63293715
|
1.987
|
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A
|
|
chr12_+_93066370
|
1.981
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr10_+_124897627
|
1.976
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr1_-_29321599
|
1.975
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr15_-_63457378
|
1.969
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr11_+_113435640
|
1.959
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr14_+_89597860
|
1.953
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr3_+_12813170
|
1.949
|
NM_001162499
NM_012298
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative)
|
|
chr17_+_75366524
|
1.949
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr19_+_38979502
|
1.948
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr17_+_26742767
|
1.937
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_-_6253152
|
1.936
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr2_-_100087366
|
1.922
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr3_+_53504070
|
1.920
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr2_+_104837004
|
1.915
|
|
|
|
|
chr1_-_1465602
|
1.909
|
NM_001114748
|
C1orf70
|
chromosome 1 open reading frame 70
|
|
chr7_-_27206249
|
1.883
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr18_-_28606839
|
1.877
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr14_+_95412481
|
1.867
|
|
|
|
|
chr17_-_37586716
|
1.859
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr9_-_95757448
|
1.850
|
|
BARX1
|
BARX homeobox 1
|
|
chr4_+_995395
|
1.848
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr17_+_26742903
|
1.845
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr19_-_19233370
|
1.843
|
|
HAPLN4
|
hyaluronan and proteoglycan link protein 4
|
|
chr19_-_52666886
|
1.841
|
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr11_-_6633427
|
1.830
|
NM_003737
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr20_+_55359551
|
1.830
|
NM_003610
|
RAE1
|
RAE1 RNA export 1 homolog (S. pombe)
|
|
chr19_-_16868674
|
1.823
|
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr20_-_61755145
|
1.823
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr16_+_1323627
|
1.818
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr11_+_2422747
|
1.808
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr8_+_65656164
|
1.807
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr14_-_20562962
|
1.798
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr7_-_44152252
|
1.793
|
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr5_+_3649167
|
1.782
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr2_-_200030412
|
1.782
|
|
SATB2
|
SATB homeobox 2
|
|
chr3_-_140148671
|
1.770
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr3_-_129689394
|
1.768
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr1_+_154878498
|
1.759
|
|
BCAN
|
brevican
|
|
chr7_-_72822471
|
1.754
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr7_-_44152232
|
1.754
|
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr21_-_43369492
|
1.740
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr12_+_56291484
|
1.739
|
NM_182947
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr1_-_40903700
|
1.739
|
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr19_-_55914837
|
1.737
|
|
|
|
|
chrX_+_144707022
|
1.732
|
NM_001144003
NM_001144004
NM_001144005
NM_032539
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr19_+_447453
|
1.727
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr11_-_69343128
|
1.720
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chrX_+_128942435
|
1.717
|
|
|
|
|
chr17_+_78630793
|
1.710
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr7_-_19123765
|
1.706
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr5_-_16232896
|
1.699
|
NM_001102562
|
MARCH11
|
membrane-associated ring finger (C3HC4) 11
|
|
chr12_+_52618815
|
1.698
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr6_-_165996031
|
1.694
|
|
LOC100132188
|
LP7097
|
|
chrX_+_150095713
|
1.694
|
NM_004224
|
GPR50
|
G protein-coupled receptor 50
|
|
chr20_-_30534848
|
1.694
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr10_+_94823636
|
1.693
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr22_+_43476755
|
1.674
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr3_-_10724715
|
1.671
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr12_+_56230047
|
1.668
|
|
KIF5A
|
kinesin family member 5A
|
|
chr10_-_28074752
|
1.667
|
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr22_+_48951286
|
1.666
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr10_+_94439658
|
1.661
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr17_+_26839143
|
1.658
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr13_-_101366995
|
1.656
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr17_-_7138535
|
1.652
|
|
YBX2
|
Y box binding protein 2
|
|
chr19_-_242168
|
1.636
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr7_-_19123787
|
1.628
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr9_+_134447795
|
1.623
|
NM_020064
|
BARHL1
|
BarH-like homeobox 1
|
|
chr14_+_105024576
|
1.620
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr1_+_63561632
|
1.606
|
|
FOXD3
|
forkhead box D3
|
|
chr10_+_83625049
|
1.599
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr4_+_3264509
|
1.596
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr12_+_4889309
|
1.594
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr18_+_75256915
|
1.587
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr8_+_1699418
|
1.582
|
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr5_-_146237826
|
1.578
|
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr22_+_47350781
|
1.577
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr16_-_695719
|
1.576
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chrX_-_134883798
|
1.575
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr19_+_1397268
|
1.575
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr17_+_44429748
|
1.574
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr2_+_171280182
|
1.572
|
|
|
|
|
chr19_-_14177980
|
1.571
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr16_-_47872880
|
1.566
|
|
|
|
|
chr22_+_27798887
|
1.562
|
NM_001039570
NM_032045
|
KREMEN1
|
kringle containing transmembrane protein 1
|
|
chr14_+_105024301
|
1.558
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr19_-_13478037
|
1.558
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr1_-_99242765
|
1.557
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr1_+_21708502
|
1.556
|
|
ALPL
|
alkaline phosphatase, liver/bone/kidney
|
|
chr11_-_40271086
|
1.552
|
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr11_+_279113
|
1.546
|
NM_025092
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr22_+_43442987
|
1.545
|
NM_001017528
NM_001017529
|
PRR5
|
proline rich 5 (renal)
|
|
chr2_+_210344945
|
1.541
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr5_-_153838013
|
1.538
|
NM_004821
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr17_+_35036704
|
1.536
|
NM_032192
|
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B
|
|
chr1_+_154878344
|
1.535
|
NM_021948
NM_198427
|
BCAN
|
brevican
|
|
chr1_+_1971762
|
1.533
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|