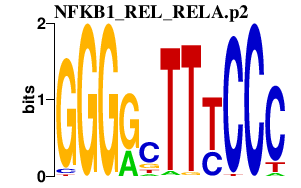
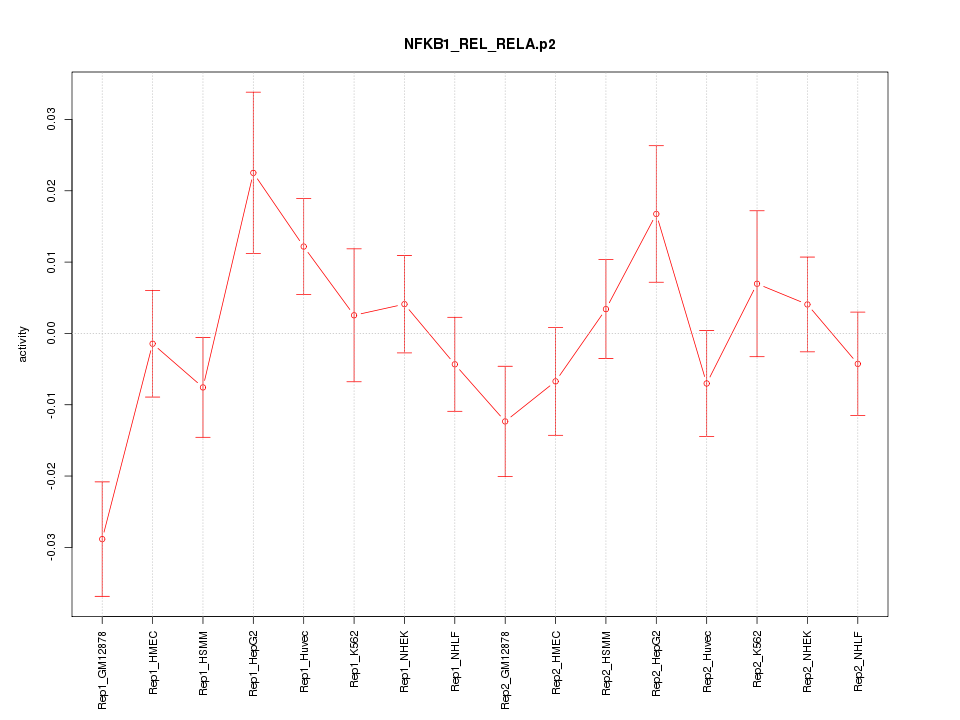
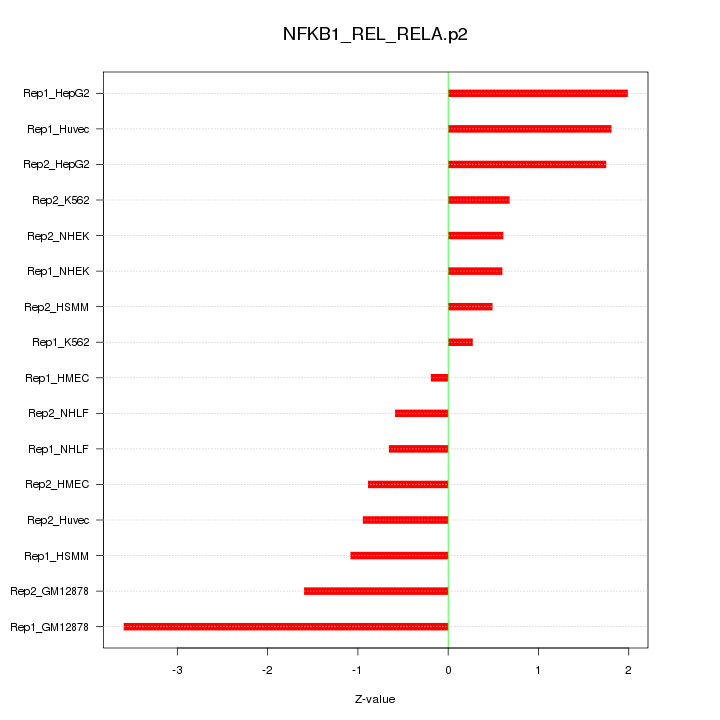
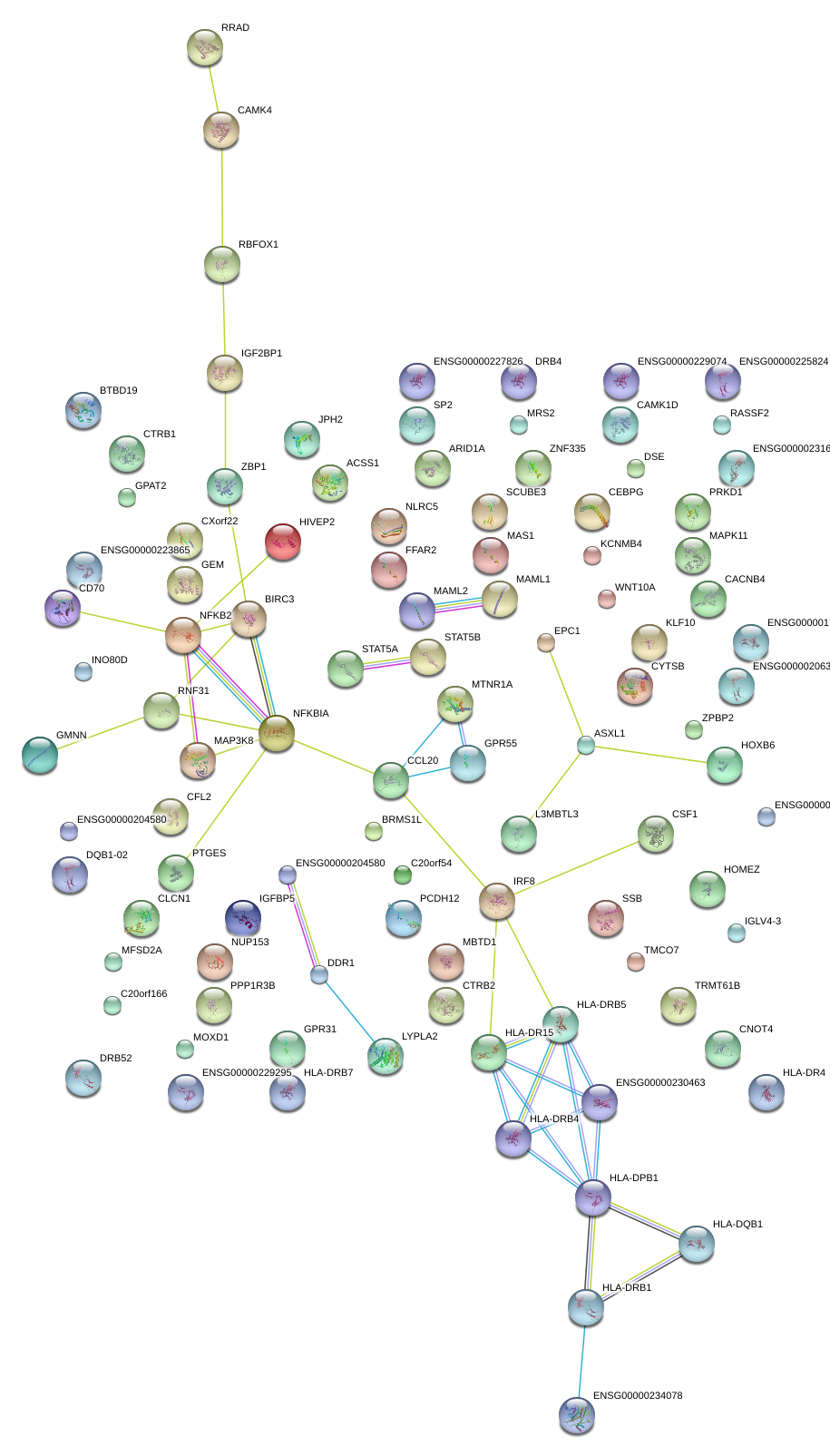
|
chr2_+_219453487
|
2.357
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr14_-_34943574
|
2.158
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr20_-_4743768
|
1.465
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr6_+_116798791
|
1.300
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr14_-_34253473
|
1.270
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_+_37694007
|
1.256
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr6_+_30958672
|
1.094
|
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr2_+_170363573
|
1.093
|
NM_003142
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr17_+_43328514
|
1.066
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr19_+_40632456
|
0.996
|
NM_005306
|
FFAR2
|
free fatty acid receptor 2
|
|
chr1_+_45046740
|
0.922
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr6_-_143307976
|
0.915
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr14_-_22825072
|
0.899
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr10_+_104145421
|
0.892
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr2_-_231498184
|
0.892
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr16_-_73798539
|
0.878
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr7_-_104440707
|
0.872
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr8_-_95343716
|
0.853
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr18_+_3584056
|
0.848
|
|
FLJ35776
|
hypothetical LOC649446
|
|
chr16_+_84490267
|
0.846
|
NM_002163
|
IRF8
|
interferon regulatory factor 8
|
|
chr1_+_110254755
|
0.840
|
NM_000757
NM_172210
NM_172211
NM_172212
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr6_-_132764257
|
0.829
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr10_-_32707549
|
0.829
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr17_+_73054614
|
0.821
|
|
|
|
|
chr5_-_141318810
|
0.808
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr20_-_24986501
|
0.804
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr6_-_17814596
|
0.799
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr10_+_12431477
|
0.771
|
NM_020397
NM_153498
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr14_+_35365266
|
0.768
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr20_-_55628852
|
0.766
|
NM_001160417
NM_001160418
NM_001160419
NM_030776
|
ZBP1
|
Z-DNA binding protein 1
|
|
chr20_+_60558369
|
0.722
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr17_-_46692369
|
0.719
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr6_+_130381408
|
0.711
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr17_+_35277942
|
0.708
|
NM_198844
NM_199321
|
ZPBP2
|
zona pellucida binding protein 2
|
|
chr17_+_19999784
|
0.708
|
NM_001033554
NM_001033555
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr2_+_233093416
|
0.703
|
NM_001195129
|
LOC646960
|
trypsin serine protease-like
|
|
chr20_-_44034194
|
0.701
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr16_+_55580910
|
0.687
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr5_+_110587546
|
0.681
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr20_-_42249146
|
0.679
|
|
JPH2
|
junctophilin 2
|
|
chr14_-_29466562
|
0.679
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr11_+_101693403
|
0.675
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr6_+_31476400
|
0.671
|
|
|
|
|
chr6_-_167491305
|
0.671
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr16_-_65516919
|
0.669
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr2_-_206659140
|
0.667
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr2_-_28946631
|
0.648
|
NM_017910
|
TRMT61B
|
tRNA methyltransferase 61 homolog B (S. cerevisiae)
|
|
chr12_+_69046241
|
0.648
|
NM_014505
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr6_+_35289699
|
0.647
|
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr6_+_24511123
|
0.638
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr2_-_96064389
|
0.621
|
NM_207328
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr8_-_103737058
|
0.618
|
NM_005655
|
KLF10
|
Kruppel-like factor 10
|
|
chr7_-_134845363
|
0.617
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr6_-_32659958
|
0.613
|
|
HLA-DRB1
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 5
|
|
chr11_-_95715976
|
0.603
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr14_+_23685935
|
0.600
|
|
RNF31
|
ring finger protein 31
|
|
chr2_+_228386800
|
0.598
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr2_-_217248967
|
0.596
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr7_+_142723340
|
0.595
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr20_-_44034346
|
0.595
|
|
ZNF335
|
zinc finger protein 335
|
|
chr20_+_30410447
|
0.592
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr9_-_131555111
|
0.586
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr8_-_9045548
|
0.585
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr22_-_49050847
|
0.585
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr4_-_187713512
|
0.580
|
NM_005958
|
MTNR1A
|
melatonin receptor 1A
|
|
chr1_+_26896519
|
0.579
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr19_+_38556414
|
0.576
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr1_+_40193424
|
0.573
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr16_+_6009095
|
0.568
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_+_23990228
|
0.567
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr19_-_6542012
|
0.562
|
|
CD70
|
CD70 molecule
|
|
chr6_+_160247962
|
0.561
|
NM_002377
|
MAS1
|
MAS1 oncogene
|
|
chr17_-_44037332
|
0.557
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr20_-_697127
|
0.555
|
NM_033409
|
C20orf54
|
chromosome 20 open reading frame 54
|
|
chr10_+_30763056
|
0.553
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr22_+_21543671
|
0.551
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr5_+_179092405
|
0.550
|
NM_014757
|
MAML1
|
mastermind-like 1 (Drosophila)
|
|
chr2_-_152663748
|
0.545
|
NM_000726
NM_001145798
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr16_+_67434993
|
0.542
|
NM_024562
|
TMCO7
|
transmembrane and coiled-coil domains 7
|
|
chr2_-_152663454
|
0.542
|
NM_001005746
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_+_68815416
|
0.539
|
NM_001007231
NM_001166277
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr11_+_18244383
|
0.538
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chr2_-_162883062
|
0.537
|
NM_022168
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr2_-_240634161
|
0.535
|
NM_173351
|
OR6B3
|
olfactory receptor, family 6, subfamily B, member 3
|
|
chr11_+_46255737
|
0.534
|
NM_052854
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1
|
|
chr20_+_762355
|
0.534
|
NM_001042353
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr20_-_42249631
|
0.533
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chr6_-_167717907
|
0.533
|
NM_004610
|
TCP10
|
t-complex 10 homolog (mouse)
|
|
chr2_-_201536425
|
0.531
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr2_-_240618518
|
0.529
|
NM_001005853
|
OR6B2
|
olfactory receptor, family 6, subfamily B, member 2
|
|
chr1_-_200397300
|
0.529
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr9_+_4480193
|
0.521
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr6_+_111911226
|
0.520
|
|
LOC643749
|
hypothetical LOC643749
|
|
chr6_-_20320607
|
0.513
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr6_+_31782618
|
0.506
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr14_+_18623364
|
0.495
|
NM_001005356
|
POTEG
|
POTE ankyrin domain family, member G
|
|
chr16_-_79668304
|
0.495
|
NM_001100873
NM_152337
|
C16orf46
|
chromosome 16 open reading frame 46
|
|
chr4_-_147662318
|
0.494
|
NM_001029998
NM_032128
|
SLC10A7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7
|
|
chr2_+_183289205
|
0.493
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr15_+_74139325
|
0.488
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr14_-_22825148
|
0.482
|
NM_020834
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr15_-_43280664
|
0.482
|
NM_138356
|
SHF
|
Src homology 2 domain containing F
|
|
chr19_+_2427122
|
0.480
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr10_-_21854616
|
0.478
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr17_-_50854054
|
0.475
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr16_-_14695930
|
0.473
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr16_-_73292188
|
0.470
|
NM_001142497
NM_152649
|
MLKL
|
mixed lineage kinase domain-like
|
|
chr2_+_44442546
|
0.467
|
NM_024766
|
C2orf34
|
chromosome 2 open reading frame 34
|
|
chr16_+_29942155
|
0.465
|
NM_001109659
NM_001109660
|
C16orf92
|
chromosome 16 open reading frame 92
|
|
chr7_-_21951946
|
0.464
|
NM_001127370
NM_001127371
NM_018719
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr1_+_165865953
|
0.460
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr4_-_113656762
|
0.459
|
NM_024019
|
NEUROG2
|
neurogenin 2
|
|
chr8_-_95343696
|
0.458
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr6_-_150080852
|
0.457
|
|
LATS1
|
LATS, large tumor suppressor, homolog 1 (Drosophila)
|
|
chr4_-_174327377
|
0.456
|
|
|
|
|
chr14_-_20642214
|
0.455
|
|
|
|
|
chr5_-_54866126
|
0.454
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr6_+_44352443
|
0.453
|
|
TMEM151B
|
transmembrane protein 151B
|
|
chr6_+_149109841
|
0.451
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr1_+_25629968
|
0.444
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr2_+_42249896
|
0.442
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr20_+_28225511
|
0.441
|
|
LOC283788
FRG1B
|
FSHD region gene 1 pseudogene
FSHD region gene 1 family, member B
|
|
chr3_-_130195608
|
0.440
|
NM_020741
|
KIAA1257
|
KIAA1257
|
|
chr1_-_205210567
|
0.438
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr7_+_149042960
|
0.437
|
NM_032534
|
KRBA1
|
KRAB-A domain containing 1
|
|
chr12_-_74711642
|
0.435
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr1_-_183210155
|
0.435
|
|
FAM129A
|
family with sequence similarity 129, member A
|
|
chr4_+_609362
|
0.434
|
NM_000283
NM_001145291
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta
|
|
chr1_-_1667123
|
0.429
|
NM_182838
|
SLC35E2B
SLC35E2
|
solute carrier family 35, member E2B
solute carrier family 35, member E2
|
|
chr6_+_35290160
|
0.425
|
NM_152753
|
SCUBE3
|
signal peptide, CUB domain, EGF-like 3
|
|
chr18_-_42804291
|
0.423
|
NM_001100817
|
TCEB3C
TCEB3CL
|
transcription elongation factor B polypeptide 3C (elongin A3)
transcription elongation factor B polypeptide 3C-like
|
|
chr17_-_17435631
|
0.420
|
NM_148172
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_+_16220952
|
0.420
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr6_+_14225932
|
0.420
|
|
CD83
|
CD83 molecule
|
|
chr10_-_51293220
|
0.419
|
NM_006327
|
TIMM23
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog (yeast)
translocase of inner mitochondrial membrane 23 homolog B (yeast)
|
|
chr20_-_32163576
|
0.415
|
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr22_+_21094094
|
0.413
|
|
|
|
|
chr11_+_1846478
|
0.412
|
NM_001013253
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr8_-_120914233
|
0.411
|
NM_003184
|
TAF2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa
|
|
chr2_-_196744546
|
0.411
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr2_+_115635982
|
0.408
|
NM_001178034
NM_001004360
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr2_-_88707978
|
0.408
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr2_-_20886293
|
0.404
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr6_+_31648071
|
0.403
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr13_+_113369470
|
0.401
|
NM_002929
|
GRK1
|
G protein-coupled receptor kinase 1
|
|
chr1_-_202730135
|
0.400
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr12_+_92595281
|
0.400
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr6_+_135544138
|
0.399
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr17_-_59172897
|
0.399
|
NM_001003786
NM_001003787
NM_001003788
NM_001165969
NM_001165970
NM_153335
|
STRADA
|
STE20-related kinase adaptor alpha
|
|
chr16_+_10945815
|
0.397
|
NM_015226
|
CLEC16A
|
C-type lectin domain family 16, member A
|
|
chr17_-_59172852
|
0.396
|
|
STRADA
|
STE20-related kinase adaptor alpha
|
|
chr1_+_144188375
|
0.395
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr8_+_38877909
|
0.392
|
NM_021623
|
PLEKHA2
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2
|
|
chr14_-_23878871
|
0.391
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr14_-_73323629
|
0.389
|
NM_001043318
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr14_-_21074875
|
0.389
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr14_-_23685629
|
0.387
|
NM_002818
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr19_-_52426055
|
0.387
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr3_+_120496180
|
0.386
|
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr2_-_152664056
|
0.385
|
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr12_+_68923477
|
0.384
|
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr14_-_19090251
|
0.384
|
NM_001145442
|
POTEG
POTEM
|
POTE ankyrin domain family, member G
POTE ankyrin domain family, member M
|
|
chr3_-_126576693
|
0.383
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr15_+_42507123
|
0.383
|
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr17_-_76720342
|
0.381
|
|
AATK
|
apoptosis-associated tyrosine kinase
|
|
chr1_+_159959072
|
0.379
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr22_-_14667916
|
0.379
|
NM_001136213
|
POTEH
POTEG
|
POTE ankyrin domain family, member H
POTE ankyrin domain family, member G
|
|
chr17_+_35718948
|
0.377
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr8_-_70909754
|
0.377
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr5_-_55326358
|
0.376
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr20_+_44070946
|
0.376
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr2_+_238432925
|
0.374
|
NM_005855
|
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1
|
|
chr1_+_28752106
|
0.373
|
NM_017846
|
TRNAU1AP
|
tRNA selenocysteine 1 associated protein 1
|
|
chr12_-_55310261
|
0.373
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr10_+_12431714
|
0.373
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr14_+_22911857
|
0.371
|
NM_172314
NM_022789
|
IL25
|
interleukin 25
|
|
chr11_+_120828275
|
0.370
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr14_+_20283938
|
0.370
|
NM_006683
|
EDDM3A
|
epididymal protein 3A
|
|
chr1_-_32574185
|
0.370
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr17_+_75776193
|
0.368
|
NM_052819
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr14_+_23908141
|
0.366
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr20_+_32926452
|
0.366
|
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2
|
|
chr2_-_128155829
|
0.364
|
NM_001161404
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr15_+_83724743
|
0.363
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr17_+_52026058
|
0.360
|
NM_005450
|
NOG
|
noggin
|
|
chr11_-_57054756
|
0.356
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr6_+_31647854
|
0.356
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr7_-_110989582
|
0.356
|
NM_032549
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr1_-_85235210
|
0.355
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr16_+_83239627
|
0.354
|
NM_024731
|
KLHL36
|
kelch-like 36 (Drosophila)
|
|
chr20_+_43943254
|
0.354
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr10_+_49184658
|
0.351
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr19_+_54786723
|
0.349
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr2_-_127679812
|
0.348
|
NM_001001665
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr15_-_62125562
|
0.348
|
NM_014326
|
DAPK2
|
death-associated protein kinase 2
|
|
chr3_+_171558215
|
0.344
|
|
SKIL
|
SKI-like oncogene
|
|
chr11_+_35596310
|
0.344
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr10_-_61139328
|
0.343
|
NM_194298
|
LOC100129721
SLC16A9
|
hypothetical protein LOC100129721
solute carrier family 16, member 9 (monocarboxylic acid transporter 9)
|
|
chr14_+_70444191
|
0.343
|
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chr17_+_4795128
|
0.342
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|