|
chr6_+_26232351
|
1.275
|
NM_003512
|
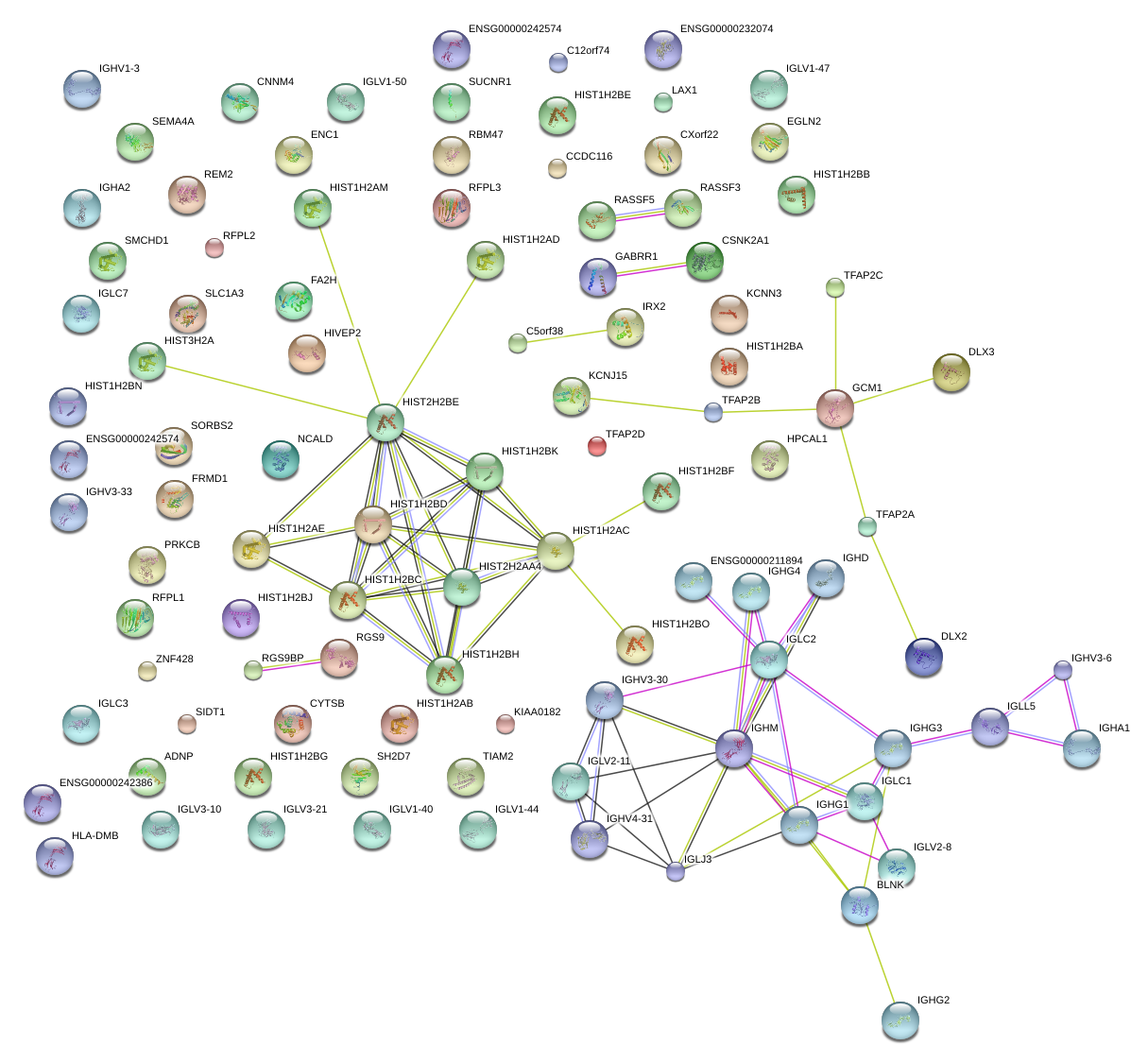
HIST1H2AC
|
histone cluster 1, H2ac
|
|
chr6_-_33016561
|
1.114
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr22_+_21407066
|
0.989
|
|
|
|
|
chr22_+_21370394
|
0.917
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr6_-_26232109
|
0.825
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr6_-_10523203
|
0.820
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr6_+_26325124
|
0.812
|
NM_021052
|
HIST1H2AE
|
histone cluster 1, H2ae
|
|
chr6_+_26359805
|
0.794
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr22_+_21495273
|
0.780
|
|
|
|
|
chr17_+_60563963
|
0.731
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr6_+_26266311
|
0.708
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr6_-_10523424
|
0.705
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr17_+_19999784
|
0.705
|
NM_001033554
NM_001033555
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr6_-_168222645
|
0.679
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr22_+_21094094
|
0.676
|
|
|
|
|
chr11_-_11331411
|
0.659
|
|
CSNK2A1P
|
casein kinase 2, alpha 1 polypeptide pseudogene
|
|
chr1_+_154386358
|
0.651
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr6_-_27222556
|
0.650
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr22_+_21116283
|
0.641
|
|
|
|
|
chr1_-_153098811
|
0.640
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_-_73365975
|
0.639
|
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr22_+_31080871
|
0.625
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chr22_+_20899155
|
0.619
|
|
|
|
|
chr5_+_2805261
|
0.619
|
NM_178569
|
C5orf38
|
chromosome 5 open reading frame 38
|
|
chr6_+_155579788
|
0.617
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr14_+_22422270
|
0.611
|
NM_173527
|
REM2
|
RAS (RAD and GEM)-like GTP binding 2
|
|
chr22_+_20316968
|
0.607
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr6_-_89984214
|
0.581
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr1_+_202000906
|
0.576
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr22_+_21042091
|
0.569
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr22_+_21340757
|
0.568
|
|
|
|
|
chr6_-_26324850
|
0.568
|
NM_003518
|
HIST1H2BG
NCALD
|
histone cluster 1, H2bg
neurocalcin delta
|
|
chr16_-_73366200
|
0.564
|
NM_024306
|
FA2H
|
fatty acid 2-hydroxylase
|
|
chr10_-_98021144
|
0.538
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr15_+_76171981
|
0.537
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr16_+_84202515
|
0.531
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr17_+_60563917
|
0.527
|
NM_001081955
NM_001165933
NM_003835
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr19_+_45996881
|
0.523
|
NM_080732
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr6_-_143307976
|
0.513
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr12_+_91620749
|
0.510
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr6_+_27208763
|
0.504
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr2_+_96790298
|
0.504
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr5_-_73972055
|
0.491
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr6_-_53121540
|
0.485
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr21_+_38566298
|
0.480
|
NM_170737
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr6_+_25834983
|
0.477
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr22_+_21384862
|
0.477
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr5_-_2804768
|
0.467
|
NM_001134222
NM_033267
|
IRX2
|
iroquois homeobox 2
|
|
chr4_-_186970366
|
0.463
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr19_+_45996940
|
0.457
|
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr3_+_153074118
|
0.455
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr19_-_48815853
|
0.446
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr6_+_26291936
|
0.444
|
NM_003523
|
HIST1H2BE
|
histone cluster 1, H2be
|
|
chr22_+_20715340
|
0.441
|
|
|
|
|
chr16_+_23754822
|
0.440
|
|
PRKCB
|
protein kinase C, beta
|
|
chr4_-_40212670
|
0.438
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr17_-_45427565
|
0.437
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr22_+_21006814
|
0.421
|
|
|
|
|
chr16_+_23754860
|
0.413
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_204797079
|
0.411
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr14_-_105862080
|
0.410
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr6_-_27208507
|
0.404
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr6_-_56515640
|
0.403
|
|
|
|
|
chr20_-_48980858
|
0.400
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr3_+_114733907
|
0.399
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr5_+_36644187
|
0.398
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_42738535
|
0.396
|
|
TMSB4XP1
|
thymosin beta 4, X-linked pseudogene 1
|
|
chr9_-_126573349
|
0.394
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr6_+_34312906
|
0.393
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_+_20880159
|
0.392
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr12_+_14409875
|
0.392
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr6_+_37245860
|
0.389
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr6_-_11887265
|
0.380
|
NM_001143948
NM_032744
|
C6orf105
|
chromosome 6 open reading frame 105
|
|
chr14_-_23623657
|
0.379
|
NM_006177
|
NRL
|
neural retina leucine zipper
|
|
chr6_-_170442090
|
0.378
|
|
|
|
|
chr1_+_165564904
|
0.378
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr22_+_20880112
|
0.378
|
|
|
|
|
chr6_+_34312667
|
0.370
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_+_165565006
|
0.369
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_-_25834733
|
0.364
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr19_+_40541327
|
0.363
|
NM_005304
|
FFAR3
|
free fatty acid receptor 3
|
|
chr16_+_23754997
|
0.362
|
|
PRKCB
|
protein kinase C, beta
|
|
chr7_+_111908143
|
0.359
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr2_-_113310796
|
0.356
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr8_+_56954953
|
0.355
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr12_-_54646061
|
0.348
|
NM_006928
|
PMEL
|
premelanosome protein
|
|
chr21_+_29322100
|
0.344
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr13_-_35327798
|
0.341
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr1_-_2447848
|
0.337
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr14_-_19252330
|
0.331
|
NM_001197287
|
OR11H2
|
olfactory receptor, family 11, subfamily H, member 2
|
|
chr3_+_122794859
|
0.331
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr6_-_26141774
|
0.327
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr15_+_78520634
|
0.322
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr6_+_20510019
|
0.318
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr1_+_204797120
|
0.315
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr17_-_53849929
|
0.306
|
NM_017763
|
RNF43
|
ring finger protein 43
|
|
chr3_+_99733432
|
0.306
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr16_+_23754772
|
0.304
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr17_+_54997667
|
0.303
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr3_+_187917791
|
0.301
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr22_+_21543671
|
0.300
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr16_-_57309625
|
0.300
|
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr6_+_27969181
|
0.294
|
NM_003527
|
HIST1H2BO
|
histone cluster 1, H2bo
|
|
chr6_+_34313007
|
0.292
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr20_+_32755778
|
0.290
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr10_+_106103503
|
0.288
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr1_+_32311084
|
0.286
|
NM_018056
|
TMEM39B
|
transmembrane protein 39B
|
|
chr1_+_32311108
|
0.286
|
|
TMEM39B
|
transmembrane protein 39B
|
|
chr6_+_53043729
|
0.284
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr16_-_87280380
|
0.282
|
NM_178310
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr7_-_112514990
|
0.282
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr10_-_62002672
|
0.282
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr22_+_21065134
|
0.281
|
|
LOC100290481
|
immunoglobulin lambda light chain-like
|
|
chr17_-_16816113
|
0.280
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr17_-_53849888
|
0.278
|
|
RNF43
|
ring finger protein 43
|
|
chr6_+_34312979
|
0.277
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr6_-_128263868
|
0.273
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr17_-_34575939
|
0.271
|
NM_001143968
|
ARL5C
|
ADP-ribosylation factor-like 5C
|
|
chr6_+_27222839
|
0.263
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr17_+_24394281
|
0.260
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr5_+_73145098
|
0.257
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr1_+_200246370
|
0.257
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr7_-_112515068
|
0.257
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr5_-_66528330
|
0.255
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr11_-_34336102
|
0.252
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr8_+_56954946
|
0.251
|
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr1_-_6537139
|
0.251
|
|
NOL9
|
nucleolar protein 9
|
|
chr22_+_21065202
|
0.250
|
|
|
|
|
chr22_+_21065207
|
0.249
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr6_+_139136349
|
0.244
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr2_-_206790871
|
0.244
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr7_-_112515035
|
0.242
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr8_+_56954899
|
0.237
|
NM_001111097
NM_002350
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
|
|
chr16_-_87280289
|
0.236
|
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr1_+_200246312
|
0.236
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr9_+_128716867
|
0.236
|
NM_001190729
NM_001190730
NM_014636
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr12_-_47250069
|
0.236
|
NM_002289
|
LALBA
|
lactalbumin, alpha-
|
|
chr14_-_105644300
|
0.232
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr8_+_123863169
|
0.228
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr5_-_16561936
|
0.227
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_+_14554895
|
0.223
|
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr6_+_21701942
|
0.222
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr12_+_52653176
|
0.220
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr1_-_181889070
|
0.219
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr9_-_94226380
|
0.219
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr12_-_11066436
|
0.217
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr20_+_62266217
|
0.217
|
|
MYT1
|
myelin transcription factor 1
|
|
chr2_-_172458970
|
0.213
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr15_+_65245546
|
0.213
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr11_-_62115545
|
0.211
|
NM_022830
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific
|
|
chr6_+_41118214
|
0.210
|
NM_001010873
NM_001159726
|
TSPO2
|
translocator protein 2
|
|
chr1_+_201711505
|
0.210
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr7_+_100021862
|
0.206
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr1_-_6537171
|
0.203
|
NM_024654
|
NOL9
|
nucleolar protein 9
|
|
chr7_-_102576570
|
0.202
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr6_-_26151863
|
0.201
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr16_+_10878539
|
0.200
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr12_-_55730125
|
0.198
|
NM_005379
|
MYO1A
|
myosin IA
|
|
chr9_+_108726257
|
0.198
|
|
ZNF462
|
zinc finger protein 462
|
|
chr19_-_18253413
|
0.198
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr12_-_31370387
|
0.194
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr4_-_91979113
|
0.193
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr19_-_45253947
|
0.190
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr10_-_64246129
|
0.190
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr6_-_11340887
|
0.188
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr20_-_60415729
|
0.187
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr14_+_18447521
|
0.187
|
NM_001013354
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12
|
|
chr7_-_27206249
|
0.186
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr2_-_175170674
|
0.183
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_-_64246080
|
0.183
|
|
EGR2
|
early growth response 2
|
|
chr12_-_55614311
|
0.182
|
NM_148897
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7
|
|
chr20_-_39680405
|
0.181
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chr4_-_170314382
|
0.180
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr14_-_33489708
|
0.180
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr5_+_140730014
|
0.179
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr6_-_170441493
|
0.179
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr17_-_38529638
|
0.179
|
NM_007298
|
BRCA1
|
breast cancer 1, early onset
|
|
chr11_+_27019084
|
0.178
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr6_+_34312641
|
0.178
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr22_-_41340886
|
0.177
|
NM_032311
NM_178136
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr6_-_11340886
|
0.176
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_64813459
|
0.175
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr17_-_44249379
|
0.175
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chrX_-_55037235
|
0.175
|
NM_002625
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
|
|
chr5_-_142045770
|
0.174
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_30783631
|
0.174
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr4_-_74307637
|
0.173
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr22_-_41340781
|
0.173
|
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3
|
|
chr6_-_11340869
|
0.171
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr3_-_52688768
|
0.169
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr8_+_123863224
|
0.168
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr1_+_201711578
|
0.167
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr22_-_22426551
|
0.167
|
NM_013378
|
VPREB3
|
pre-B lymphocyte 3
|
|
chr2_+_54638957
|
0.166
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr8_+_123863075
|
0.165
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr12_-_120725210
|
0.164
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr2_-_152393209
|
0.161
|
NM_001037174
NM_012097
NM_177985
|
ARL5A
|
ADP-ribosylation factor-like 5A
|
|
chr18_-_45271938
|
0.160
|
|
RPL17-C18ORF32
RPL17
|
RPL17-C18orf32 read-through transcript
ribosomal protein L17
|
|
chr3_-_48569213
|
0.160
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr1_-_148050534
|
0.160
|
NM_001024599
NM_001161334
|
HIST2H2BF
|
histone cluster 2, H2bf
|