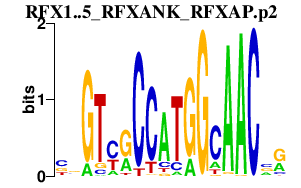
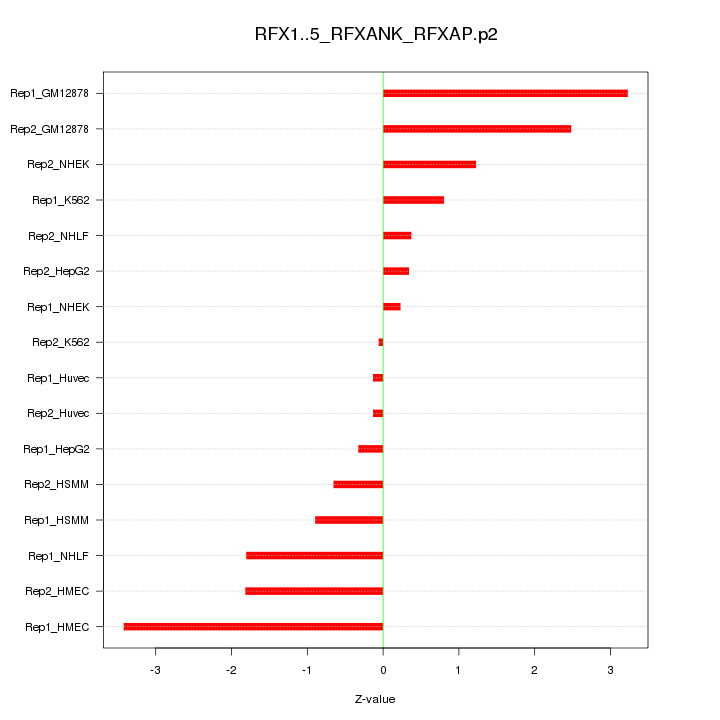
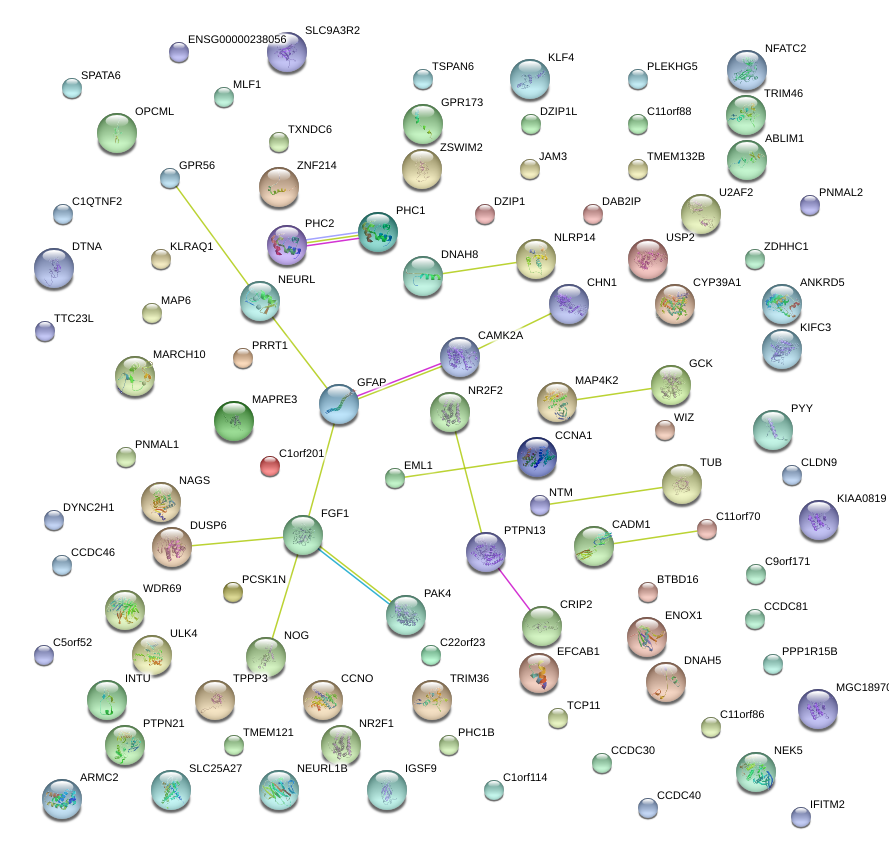
|
chr17_-_61618276
|
2.369
|
NM_145036
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr2_-_187422141
|
2.226
|
NM_182521
|
ZSWIM2
|
zinc finger, SWIM-type containing 2
|
|
chr17_-_58239382
|
2.104
|
NM_152598
|
MARCH10
|
membrane-associated ring finger (C3HC4) 10
|
|
chr11_+_93885342
|
1.812
|
NM_001190462
|
LOC643037
|
hypothetical protein LOC643037
|
|
chr2_+_27046952
|
1.651
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr1_-_33669190
|
1.430
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr14_-_88087575
|
1.395
|
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr13_-_95092283
|
1.381
|
|
DZIP1
|
DAZ interacting protein 1
|
|
chr5_-_149649593
|
1.349
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr5_+_92944680
|
1.332
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr3_-_139315011
|
1.307
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr11_+_110890719
|
1.283
|
NM_001100388
NM_207430
|
C11orf88
|
chromosome 11 open reading frame 88
|
|
chr6_+_109276280
|
1.277
|
NM_032131
|
ARMC2
|
armadillo repeat containing 2
|
|
chrX_-_48578878
|
1.246
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr1_+_42701460
|
1.191
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr19_-_51666503
|
1.142
|
|
PNMAL1
|
PNMA-like 1
|
|
chr10_-_116382089
|
1.129
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr5_-_54565181
|
1.121
|
NM_021147
|
CCNO
|
cyclin O
|
|
chr16_-_65984836
|
1.077
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr19_-_51690989
|
1.071
|
NM_020709
|
PNMAL2
|
PNMA-like 2
|
|
chr16_-_56393871
|
1.060
|
|
KIFC3
|
kinesin family member C3
|
|
chr11_+_101423378
|
1.040
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr13_-_43259032
|
1.019
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr19_-_51666629
|
1.012
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr5_-_149649380
|
1.005
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr1_-_202647526
|
0.981
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory (inhibitor) subunit 15B
|
|
chr16_+_3002457
|
0.973
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr14_+_99329198
|
0.962
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr17_+_75625025
|
0.962
|
NM_017950
|
CCDC40
|
coiled-coil domain containing 40
|
|
chr2_-_175578156
|
0.962
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr2_+_228444570
|
0.955
|
NM_178821
|
WDR69
|
WD repeat domain 69
|
|
chr16_-_56393881
|
0.936
|
NM_001130100
NM_005550
|
KIFC3
|
kinesin family member C3
|
|
chr6_-_32227695
|
0.934
|
NM_030651
|
PRRT1
|
proline-rich transmembrane protein 1
|
|
chr17_+_39437439
|
0.934
|
NM_153006
|
NAGS
|
N-acetylglutamate synthase
|
|
chr17_+_52026058
|
0.926
|
NM_005450
|
NOG
|
noggin
|
|
chr3_-_139531417
|
0.891
|
NM_178130
|
TXNDC6
|
thioredoxin domain containing 6
|
|
chr16_+_56230707
|
0.886
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chrX_+_53095230
|
0.866
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr2_+_48521411
|
0.854
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr4_+_87734489
|
0.854
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr1_-_158182006
|
0.845
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr14_+_105063975
|
0.829
|
NM_025268
|
TMEM121
|
transmembrane protein 121
|
|
chr4_+_128773529
|
0.827
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr13_-_51601214
|
0.817
|
NM_199289
|
NEK5
|
NIMA (never in mitosis gene a)-related kinase 5
|
|
chr11_-_6998041
|
0.815
|
NM_013249
|
ZNF214
|
zinc finger protein 214
|
|
chr9_+_123501186
|
0.814
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr16_-_66007650
|
0.803
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr11_+_6998275
|
0.794
|
NM_176822
|
NLRP14
|
NLR family, pyrin domain containing 14
|
|
chr9_-_109291866
|
0.790
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr5_-_14064846
|
0.773
|
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr5_-_114533448
|
0.761
|
|
TRIM36
|
tripartite motif containing 36
|
|
chr16_+_56219802
|
0.753
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr6_-_46728188
|
0.752
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr11_-_114880275
|
0.750
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr6_-_32227645
|
0.741
|
|
PRRT1
|
proline-rich transmembrane protein 1
|
|
chr1_-_6480058
|
0.728
|
NM_001042663
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr13_+_35904408
|
0.728
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr11_+_7997366
|
0.697
|
|
TUB
|
tubby homolog (mouse)
|
|
chr6_+_46728590
|
0.687
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr1_-_24612742
|
0.680
|
NM_001199013
NM_001199014
NM_178122
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr10_+_124020810
|
0.674
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chr11_-_118757574
|
0.657
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr3_-_41978464
|
0.655
|
NM_017886
|
ULK4
|
unc-51-like kinase 4 (C. elegans)
|
|
chr15_+_94674849
|
0.655
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_48710378
|
0.655
|
NM_019073
|
SPATA6
|
spermatogenesis associated 6
|
|
chr12_-_88270369
|
0.653
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr17_-_39437362
|
0.650
|
NM_004160
|
PYY
|
peptide YY
|
|
chrX_-_99778340
|
0.645
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr22_-_16887151
|
0.644
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr1_-_6480035
|
0.644
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr12_-_88270375
|
0.642
|
NM_001946
NM_022652
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr19_+_44308224
|
0.635
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr3_+_159771646
|
0.627
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr1_+_153413675
|
0.626
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr11_-_118757477
|
0.621
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_+_85763425
|
0.619
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr16_+_2016807
|
0.618
|
NM_001130012
NM_004785
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr7_-_44195405
|
0.615
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr5_+_34875025
|
0.613
|
NM_144725
|
TTC23L
|
tetratricopeptide repeat domain 23-like
|
|
chr6_-_35216981
|
0.609
|
NM_001093728
NM_018679
|
TCP11
|
t-complex 11 homolog (mouse)
|
|
chr9_-_109291575
|
0.599
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_88270128
|
0.582
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr19_+_60857227
|
0.578
|
NM_001012478
NM_007279
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2
|
|
chr1_-_48710263
|
0.572
|
|
SPATA6
|
spermatogenesis associated 6
|
|
chr9_+_134275431
|
0.570
|
NM_207417
|
C9orf171
|
chromosome 9 open reading frame 171
|
|
chr14_+_105012106
|
0.567
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr5_-_159730196
|
0.562
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chr1_-_167663278
|
0.553
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr5_+_157031138
|
0.549
|
NM_001145132
|
C5orf52
|
chromosome 5 open reading frame 52
|
|
chr3_+_184648089
|
0.542
|
|
LOC100505687
|
hypothetical LOC100505687
|
|
chr10_+_105243724
|
0.541
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr11_+_102485293
|
0.535
|
NM_001080463
NM_001377
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1
|
|
chr18_+_30544193
|
0.534
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr20_+_9963696
|
0.523
|
NM_022096
NM_198798
|
ANKRD5
|
ankyrin repeat domain 5
|
|
chr19_-_15421761
|
0.520
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr12_+_8958397
|
0.515
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr17_-_40339919
|
0.511
|
|
GFAP
|
glial fibrillary acidic protein
|
|
chr11_+_131285747
|
0.511
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr1_+_34098662
|
0.509
|
NM_145205
|
HMGB4
|
high-mobility group box 4
|
|
chr11_+_133444175
|
0.506
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr20_-_49612745
|
0.503
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr5_-_142046175
|
0.501
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_+_172000793
|
0.499
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr11_-_75056761
|
0.489
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr12_+_124377114
|
0.488
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr22_-_36679479
|
0.478
|
NM_032561
|
C22orf23
|
chromosome 22 open reading frame 23
|
|
chr8_-_49810270
|
0.475
|
NM_001142857
NM_024593
|
EFCAB1
|
EF-hand calcium binding domain 1
|
|
chr11_+_66499323
|
0.469
|
NM_001136485
|
C11orf86
|
chromosome 11 open reading frame 86
|
|
chr6_-_2787111
|
0.461
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr5_-_16795336
|
0.459
|
|
MYO10
|
myosin X
|
|
chr10_+_63092724
|
0.456
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr5_-_157031022
|
0.453
|
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr17_-_6675731
|
0.449
|
NM_053285
|
TEKT1
|
tektin 1
|
|
chr11_-_5916424
|
0.445
|
|
TRIM5
|
tripartite motif containing 5
|
|
chr6_-_43585958
|
0.445
|
|
C6orf154
|
chromosome 6 open reading frame 154
|
|
chr20_-_49612574
|
0.424
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr16_+_29731058
|
0.423
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr11_-_75057299
|
0.423
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr1_-_6468108
|
0.420
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr21_+_45532377
|
0.418
|
|
LOC642852
|
hypothetical LOC642852
|
|
chr7_+_129919157
|
0.416
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr12_-_25597444
|
0.416
|
NM_152590
NM_001145728
NM_001145729
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr6_+_143423656
|
0.413
|
NM_016108
|
AIG1
|
androgen-induced 1
|
|
chr6_-_2787068
|
0.409
|
NM_030666
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr7_+_129919134
|
0.408
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chrX_-_13957879
|
0.404
|
NM_001042479
NM_001042480
NM_017856
|
GEMIN8
|
gem (nuclear organelle) associated protein 8
|
|
chr17_-_71648965
|
0.399
|
NM_001454
|
FOXJ1
|
forkhead box J1
|
|
chr8_-_133756943
|
0.398
|
NM_012472
|
LRRC6
|
leucine rich repeat containing 6
|
|
chr20_-_3710060
|
0.397
|
NM_015417
|
SPEF1
|
sperm flagellar 1
|
|
chr3_-_171013000
|
0.393
|
|
LRRC34
|
leucine rich repeat containing 34
|
|
chr4_-_177353707
|
0.387
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr1_+_33319287
|
0.386
|
NM_052998
|
ADC
|
arginine decarboxylase
|
|
chr20_+_42593849
|
0.384
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr22_+_21742364
|
0.384
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr6_+_117044334
|
0.383
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chrX_-_71268475
|
0.380
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr4_+_3264509
|
0.379
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_-_176273680
|
0.378
|
|
LOC730102
|
quinone oxidoreductase-like protein 2 pseudogene
|
|
chr1_-_43509187
|
0.377
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr2_-_201930311
|
0.376
|
NM_001127391
NM_139163
|
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12
|
|
chr9_-_35818728
|
0.376
|
NM_001012446
|
C9orf128
|
chromosome 9 open reading frame 128
|
|
chr3_+_38055699
|
0.374
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr7_-_157054403
|
0.373
|
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr11_+_45899747
|
0.370
|
NM_152312
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr15_+_73803351
|
0.363
|
NM_175881
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr1_-_171905593
|
0.362
|
NM_198493
|
ANKRD45
|
ankyrin repeat domain 45
|
|
chr3_+_156280129
|
0.362
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr7_+_30777512
|
0.355
|
NM_032222
|
FAM188B
|
family with sequence similarity 188, member B
|
|
chr10_-_123343320
|
0.351
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr6_+_143423738
|
0.350
|
|
AIG1
|
androgen-induced 1
|
|
chr1_-_158136521
|
0.349
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr5_-_142045770
|
0.349
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_39211706
|
0.347
|
NM_004090
|
DUSP3
|
dual specificity phosphatase 3
|
|
chr6_+_143423723
|
0.343
|
|
AIG1
|
androgen-induced 1
|
|
chr1_+_15608974
|
0.343
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr22_+_21742727
|
0.343
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr2_-_26717713
|
0.343
|
NM_001029881
|
CIB4
|
calcium and integrin binding family member 4
|
|
chrX_-_150893752
|
0.342
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr7_+_129919330
|
0.341
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr4_-_171161486
|
0.341
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr1_+_201862845
|
0.337
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr3_-_71860958
|
0.335
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr11_-_10519265
|
0.331
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr22_-_41815277
|
0.327
|
NM_012263
|
TTLL1
|
tubulin tyrosine ligase-like family, member 1
|
|
chr12_+_50104859
|
0.326
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr5_-_1165106
|
0.325
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr11_+_31347947
|
0.324
|
NM_181706
|
DNAJC24
|
DnaJ (Hsp40) homolog, subfamily C, member 24
|
|
chr2_-_130619038
|
0.322
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr1_+_207926172
|
0.319
|
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr6_-_76051351
|
0.319
|
NM_001143958
NM_018247
|
TMEM30A
|
transmembrane protein 30A
|
|
chr2_-_182229977
|
0.319
|
NM_001030311
NM_001030312
NM_001030313
NM_001160277
NM_201548
|
CERKL
|
ceramide kinase-like
|
|
chr12_-_62348506
|
0.318
|
NM_173812
|
DPY19L2
|
dpy-19-like 2 (C. elegans)
|
|
chr19_-_63775901
|
0.316
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr3_+_151171755
|
0.315
|
|
LOC646903
|
hypothetical LOC646903
|
|
chr17_-_959007
|
0.313
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr1_+_15609009
|
0.311
|
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr1_+_201711578
|
0.310
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr10_-_25344871
|
0.308
|
NM_145010
|
ENKUR
|
enkurin, TRPC channel interacting protein
|
|
chr7_+_156126157
|
0.308
|
NM_030936
NM_001184996
|
RNF32
|
ring finger protein 32
|
|
chr15_+_96304911
|
0.308
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr6_+_125346208
|
0.306
|
NM_152553
|
RNF217
|
ring finger protein 217
|
|
chr10_-_127361583
|
0.305
|
NM_001128202
|
C10orf122
|
chromosome 10 open reading frame 122
|
|
chr11_+_93394025
|
0.300
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr17_+_65612589
|
0.300
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr20_-_36322587
|
0.299
|
NM_001029864
|
KIAA1755
|
KIAA1755
|
|
chr1_+_227473458
|
0.299
|
NM_004578
|
RAB4A
|
RAB4A, member RAS oncogene family
|
|
chr7_+_48041632
|
0.298
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr18_-_45630154
|
0.297
|
|
MYO5B
|
myosin VB
|
|
chr19_-_63776442
|
0.290
|
|
MZF1
|
myeloid zinc finger 1
|
|
chr21_+_17807167
|
0.289
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr17_+_4683280
|
0.289
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr1_+_201711505
|
0.286
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr12_+_32546243
|
0.285
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr21_-_42789369
|
0.283
|
NM_080860
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr9_+_115970599
|
0.283
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr22_+_19651446
|
0.281
|
NM_001146288
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr11_-_31347896
|
0.280
|
NM_181807
|
DCDC1
|
doublecortin domain containing 1
|
|
chr11_-_75057062
|
0.278
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chrX_+_117841761
|
0.277
|
NM_173798
|
ZCCHC12
|
zinc finger, CCHC domain containing 12
|
|
chr7_+_101861007
|
0.276
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|