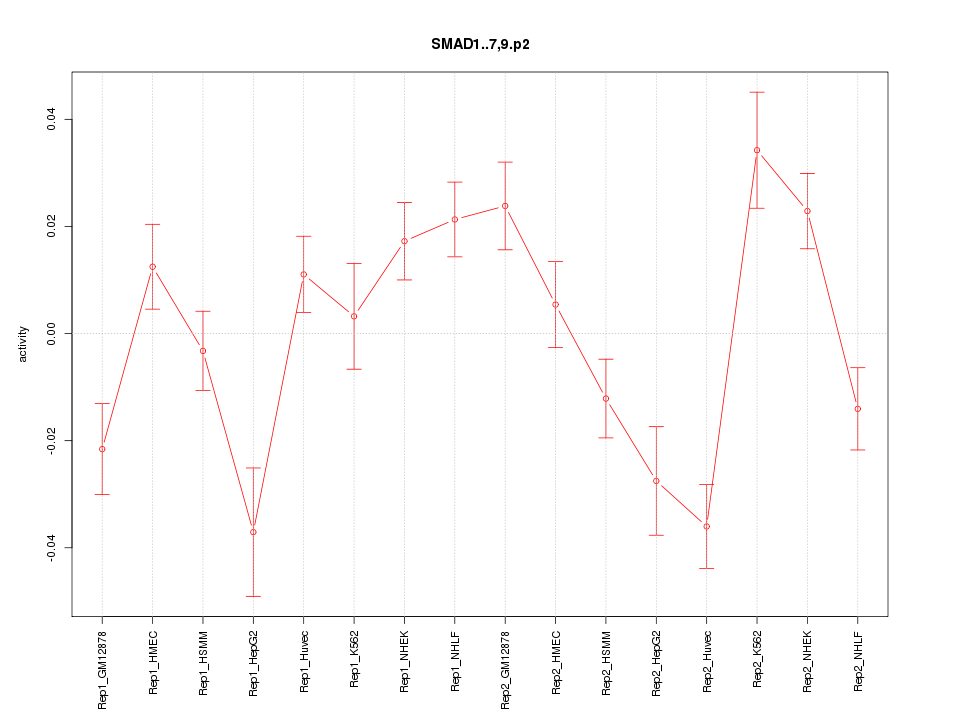
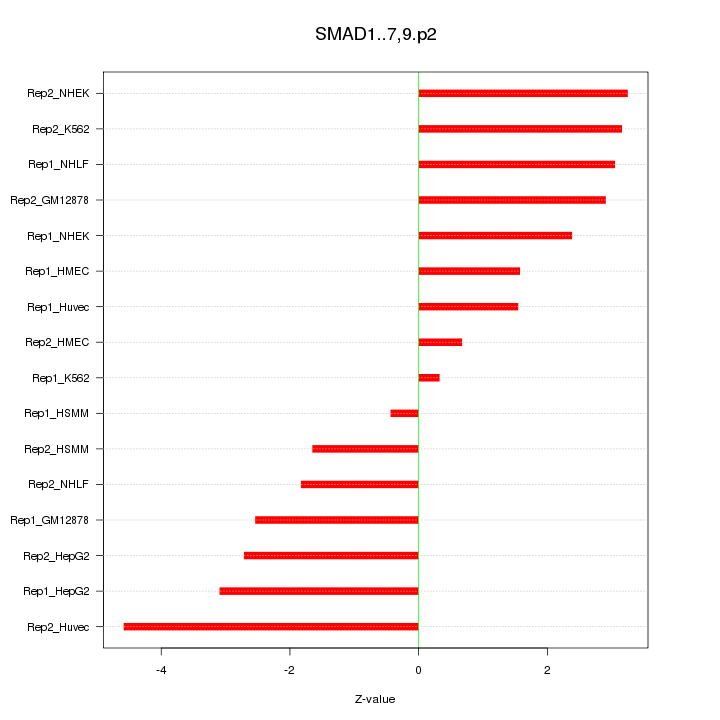
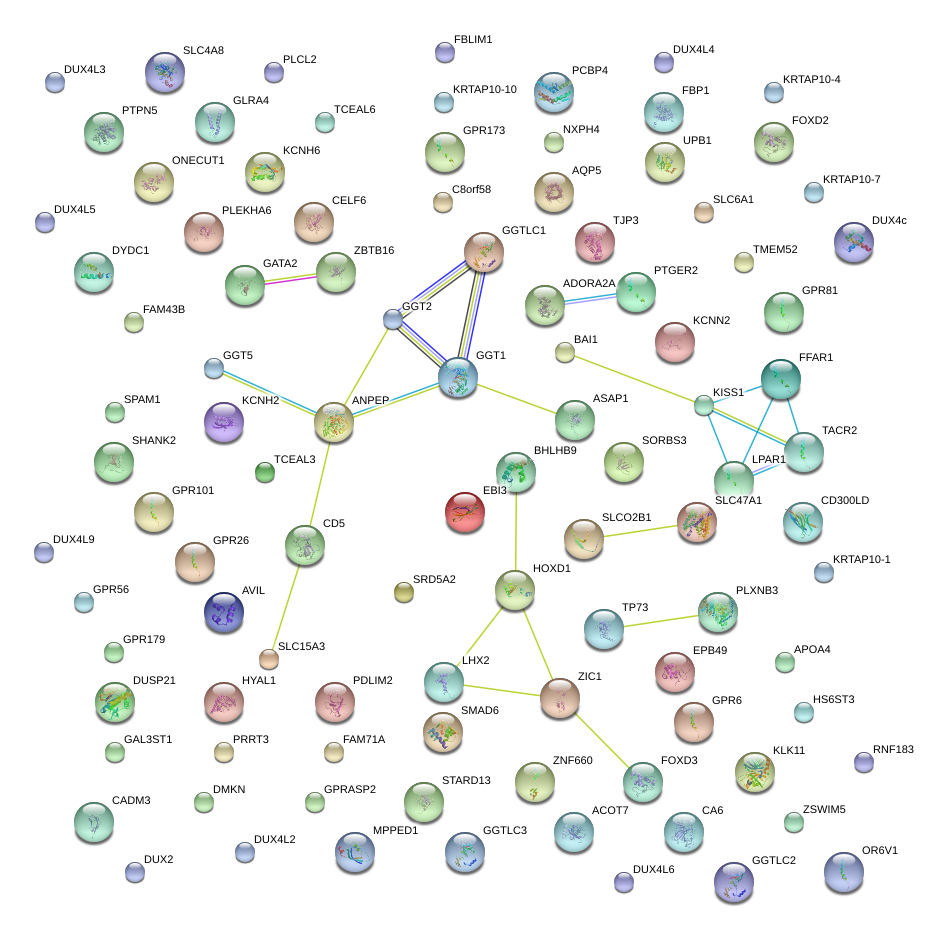
|
chr11_-_60475558
|
3.230
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr22_+_23221114
|
2.523
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr17_+_19377732
|
2.389
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr10_+_135333667
|
2.027
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191229360
|
2.001
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191242540
|
1.978
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr11_+_60626505
|
1.948
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr10_+_135330357
|
1.923
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_+_20751518
|
1.860
|
NM_207334
|
FAM43B
|
family with sequence similarity 43, member B
|
|
chr9_-_139183675
|
1.842
|
|
|
|
|
chr10_+_135343649
|
1.758
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr3_+_44601457
|
1.755
|
NM_173658
|
ZNF660
|
zinc finger protein 660
|
|
chrX_+_102749489
|
1.718
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr22_-_22970850
|
1.714
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr17_-_33753218
|
1.709
|
NM_001004334
|
GPR179
|
G protein-coupled receptor 179
|
|
chr4_+_191232653
|
1.694
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr5_+_113725914
|
1.667
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr11_-_18769703
|
1.649
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr3_-_129689394
|
1.628
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr16_+_56219802
|
1.620
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr9_-_96441654
|
1.593
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr8_+_143542378
|
1.581
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|
|
chr21_+_44844924
|
1.580
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr21_+_44818033
|
1.575
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr4_+_191226073
|
1.574
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr11_+_113436438
|
1.560
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr13_-_32822759
|
1.552
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr8_+_22465368
|
1.502
|
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr22_+_21318779
|
1.496
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr1_-_45444812
|
1.456
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr1_+_15964045
|
1.450
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr2_+_176761552
|
1.439
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chrX_-_102870207
|
1.420
|
NM_001024452
NM_001172285
|
GLRA4
|
glycine receptor, alpha 4
|
|
chrX_+_53095230
|
1.420
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr15_-_50869379
|
1.400
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chrX_+_44588192
|
1.388
|
NM_022076
|
DUSP21
|
dual specificity phosphatase 21
|
|
chr1_-_6368324
|
1.383
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr4_+_191239247
|
1.374
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr21_+_44881700
|
1.333
|
NM_181688
|
KRTAP10-10
|
keratin associated protein 10-10
|
|
chr15_-_70385709
|
1.332
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_-_202595666
|
1.318
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr9_-_96441567
|
1.316
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr3_+_148610516
|
1.316
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr7_-_150283794
|
1.306
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr1_+_8928508
|
1.302
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chr1_+_3558958
|
1.294
|
NM_005427
|
TP73
|
tumor protein p73
|
|
chrX_+_101862297
|
1.287
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr14_+_51850754
|
1.282
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr4_+_191235959
|
1.277
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr17_-_70099964
|
1.260
|
NM_001115152
|
CD300LD
|
CD300 molecule-like family member d
|
|
chr15_-_88143721
|
1.253
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr15_+_64782468
|
1.234
|
|
SMAD6
|
SMAD family member 6
|
|
chr1_+_63561632
|
1.218
|
|
FOXD3
|
forkhead box D3
|
|
chrX_+_152682772
|
1.209
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr3_-_51968995
|
1.188
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr3_+_16901455
|
1.183
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr8_+_22493928
|
1.173
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr22_+_23153529
|
1.157
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr5_+_113726203
|
1.154
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr12_+_48641545
|
1.150
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr11_-_70350292
|
1.125
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr10_+_125415860
|
1.110
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chr10_-_70846573
|
1.107
|
NM_001057
|
TACR2
|
tachykinin receptor 2
|
|
chr3_+_11009419
|
1.105
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr9_-_96442351
|
1.092
|
NM_001127628
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr22_-_29290875
|
1.092
|
NM_004861
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr1_+_210864411
|
1.084
|
NM_153606
|
FAM71A
|
family with sequence similarity 71, member A
|
|
chr21_-_44784499
|
1.050
|
NM_198691
|
KRTAP10-1
|
keratin associated protein 10-1
|
|
chr1_+_157408000
|
1.042
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr13_+_95541093
|
1.041
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr11_-_70350509
|
1.039
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr10_-_82106479
|
1.032
|
NM_138812
|
DYDC1
|
DPY30 domain containing 1
|
|
chr9_-_96442377
|
1.031
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr9_+_125813849
|
1.028
|
|
LHX2
|
LIM homeobox 2
|
|
chr11_+_74548472
|
1.024
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr19_+_3659358
|
1.010
|
|
TJP3
|
tight junction protein 3 (zona occludens 3)
|
|
chr12_-_121780835
|
1.009
|
NM_032554
|
GPR81
|
G protein-coupled receptor 81
|
|
chr1_-_202432165
|
1.008
|
NM_002256
|
KISS1
|
KiSS-1 metastasis-suppressor
|
|
chr8_+_21971317
|
1.007
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr7_+_142459538
|
1.001
|
NM_001001667
|
OR6V1
|
olfactory receptor, family 6, subfamily V, member 1
|
|
chr19_-_40684638
|
0.997
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr6_+_110406890
|
0.995
|
NM_005284
|
GPR6
|
G protein-coupled receptor 6
|
|
chr12_-_56496107
|
0.986
|
NM_006576
|
AVIL
|
advillin
|
|
chr8_-_131483395
|
0.983
|
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr20_-_23915431
|
0.982
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr9_-_115101140
|
0.975
|
NM_145051
|
RNF183
|
ring finger protein 183
|
|
chr11_-_116199145
|
0.971
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr19_+_40534284
|
0.964
|
NM_005303
|
FFAR1
|
free fatty acid receptor 1
|
|
chrX_+_101853912
|
0.950
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr12_+_50104936
|
0.945
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr9_-_96441409
|
0.939
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr19_-_56221681
|
0.936
|
NM_001136032
|
KLK11
|
kallikrein-related peptidase 11
|
|
chr3_-_50324815
|
0.926
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr22_+_42137955
|
0.922
|
NM_001044370
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr2_-_31659472
|
0.917
|
NM_000348
|
SRD5A2
|
steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2)
|
|
chr1_-_1840564
|
0.909
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr12_+_55896844
|
0.907
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chrX_-_135941498
|
0.905
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr19_+_4180494
|
0.901
|
NM_005755
|
EBI3
|
Epstein-Barr virus induced 3
|
|
chr1_-_31618405
|
0.900
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr11_-_17522430
|
0.894
|
NM_005709
NM_153676
|
USH1C
|
Usher syndrome 1C (autosomal recessive, severe)
|
|
chr19_-_46626474
|
0.893
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr19_+_9064854
|
0.889
|
NM_001004456
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1
|
|
chr14_+_99273751
|
0.886
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr17_-_42411587
|
0.886
|
NM_203400
|
RPRML
|
reprimo-like
|
|
chr9_+_128764324
|
0.886
|
NM_001190728
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chrX_+_71318250
|
0.877
|
NM_001170747
NM_006223
|
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin)
|
|
chrX_-_114703273
|
0.875
|
|
|
|
|
chr4_+_55218663
|
0.870
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr1_-_164402575
|
0.864
|
NM_001017961
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr1_+_63561317
|
0.859
|
NM_012183
|
FOXD3
|
forkhead box D3
|
|
chr19_-_41034734
|
0.858
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr7_-_44195405
|
0.857
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr1_-_154913712
|
0.857
|
NM_006617
|
NES
|
nestin
|
|
chr10_+_82106537
|
0.853
|
NM_032372
|
DYDC2
|
DPY30 domain containing 2
|
|
chr22_+_42138034
|
0.853
|
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr19_+_40465249
|
0.852
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr11_+_47564805
|
0.843
|
NM_001164379
|
FAM180B
|
family with sequence similarity 180, member B
|
|
chr8_-_143820825
|
0.842
|
NM_020427
|
SLURP1
|
secreted LY6/PLAUR domain containing 1
|
|
chr7_+_154493478
|
0.833
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr6_-_25834733
|
0.830
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr21_-_44803049
|
0.828
|
NM_198696
|
KRTAP10-3
|
keratin associated protein 10-3
|
|
chr11_+_122901543
|
0.822
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr12_+_50104859
|
0.821
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr19_-_16145285
|
0.820
|
NM_054113
|
CIB3
|
calcium and integrin binding family member 3
|
|
chr5_+_156628929
|
0.819
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr1_+_24713396
|
0.817
|
|
RCAN3
|
RCAN family member 3
|
|
chr1_-_218168548
|
0.816
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr17_-_24357014
|
0.815
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr16_-_3783475
|
0.811
|
|
CREBBP
|
CREB binding protein
|
|
chr19_+_2200112
|
0.809
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr6_-_26038817
|
0.806
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chrX_-_125127760
|
0.802
|
NM_001013628
|
DCAF12L2
|
DDB1 and CUL4 associated factor 12-like 2
|
|
chr4_+_9392355
|
0.797
|
NM_000798
|
DRD5
|
dopamine receptor D5
|
|
chrX_-_106846684
|
0.797
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_-_65860450
|
0.794
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr3_+_42702014
|
0.791
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr21_+_46370831
|
0.784
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr2_+_70981233
|
0.778
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chrX_-_62487693
|
0.776
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chrX_+_102727068
|
0.771
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr4_+_191245840
|
0.770
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_-_201321999
|
0.768
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr2_+_97717300
|
0.764
|
NM_207519
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chrX_-_108754906
|
0.764
|
NM_012282
|
KCNE1L
|
KCNE1-like
|
|
chr21_+_43462209
|
0.758
|
NM_000394
|
CRYAA
|
crystallin, alpha A
|
|
chr1_+_178466064
|
0.757
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chrX_-_128616594
|
0.752
|
NM_017413
|
APLN
|
apelin
|
|
chr3_+_46717826
|
0.751
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr22_+_18330049
|
0.750
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr16_-_47872973
|
0.750
|
|
CBLN1
|
cerebellin 1 precursor
|
|
chr19_-_53808505
|
0.748
|
NM_017708
|
FAM83E
|
family with sequence similarity 83, member E
|
|
chr2_+_71016503
|
0.744
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chrX_-_50573759
|
0.744
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr21_-_31175722
|
0.740
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr3_-_62835601
|
0.736
|
|
|
|
|
chr3_+_52325374
|
0.735
|
NM_015512
|
DNAH1
|
dynein, axonemal, heavy chain 1
|
|
chr17_-_17421050
|
0.734
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr15_-_97368546
|
0.733
|
NM_001167902
|
PGPEP1L
|
pyroglutamyl-peptidase I-like
|
|
chr11_+_1420250
|
0.730
|
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr1_+_226261374
|
0.729
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr17_+_43128422
|
0.726
|
|
|
|
|
chr22_+_21407066
|
0.724
|
|
|
|
|
chr7_-_27108826
|
0.723
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr18_-_45630154
|
0.715
|
|
MYO5B
|
myosin VB
|
|
chr4_-_673107
|
0.715
|
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr19_-_56105805
|
0.715
|
NM_004917
|
KLK4
|
kallikrein-related peptidase 4
|
|
chr7_+_128367961
|
0.714
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr7_+_75977680
|
0.714
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr21_-_30614477
|
0.711
|
NM_203405
|
KRTAP26-1
|
keratin associated protein 26-1
|
|
chr22_-_20235372
|
0.710
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr2_+_70981223
|
0.709
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr18_-_43821491
|
0.709
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr9_-_115880281
|
0.707
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr14_+_105012106
|
0.702
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr15_+_72253940
|
0.702
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr7_-_105106844
|
0.701
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chrX_-_2896285
|
0.701
|
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr1_+_226712430
|
0.698
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr16_-_29817841
|
0.696
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr7_+_73080362
|
0.695
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chrX_-_37916138
|
0.691
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr21_+_44890758
|
0.690
|
NM_198692
|
KRTAP10-11
|
keratin associated protein 10-11
|
|
chr3_-_15515382
|
0.688
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chrX_+_70438352
|
0.685
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr7_-_76667045
|
0.684
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr8_+_142501188
|
0.680
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr15_+_39918302
|
0.680
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chrX_+_49531209
|
0.678
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chrX_-_114374860
|
0.677
|
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chrX_-_12972641
|
0.677
|
NM_174901
|
FAM9C
|
family with sequence similarity 9, member C
|
|
chr7_+_70235724
|
0.676
|
NM_022479
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17
|
|
chr6_-_24466258
|
0.676
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr22_-_35835548
|
0.674
|
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr21_+_44871467
|
0.672
|
NM_198690
|
KRTAP10-9
|
keratin associated protein 10-9
|
|
chr7_+_73080404
|
0.671
|
|
ELN
|
elastin
|
|
chr1_+_199884152
|
0.671
|
|
NAV1
|
neuron navigator 1
|
|
chr9_+_125813672
|
0.669
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr19_-_11550576
|
0.669
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr11_-_60379998
|
0.668
|
NM_004778
|
GPR44
|
G protein-coupled receptor 44
|