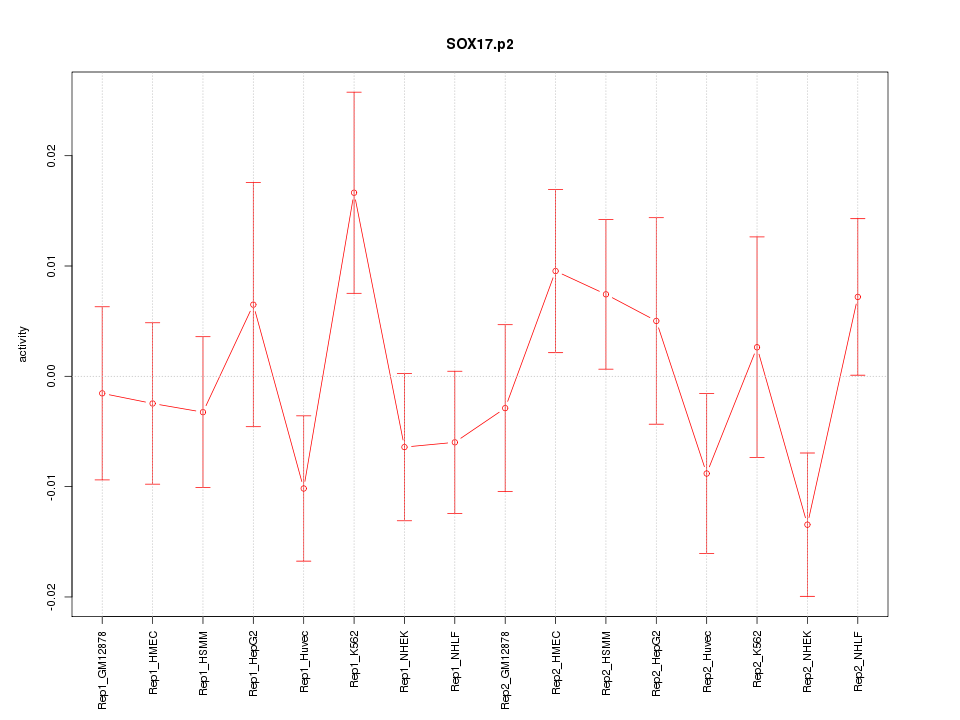
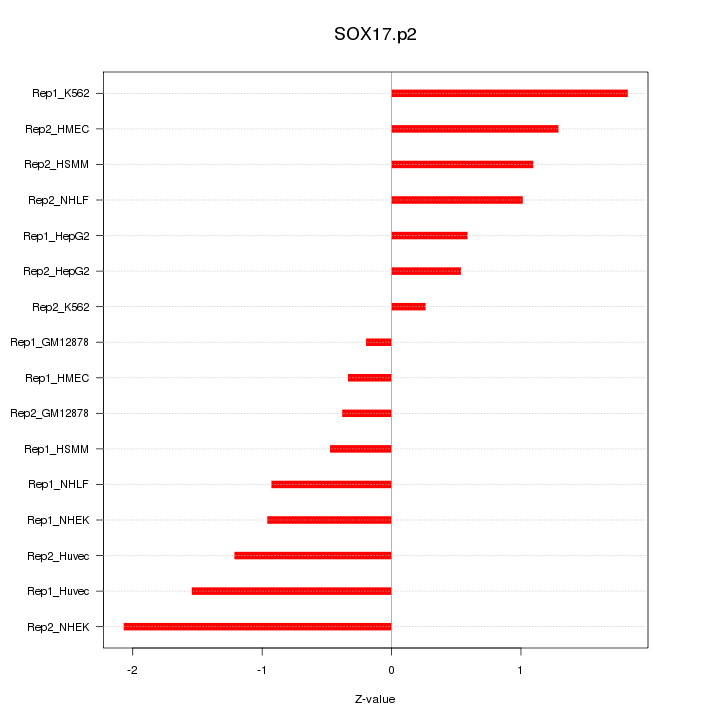
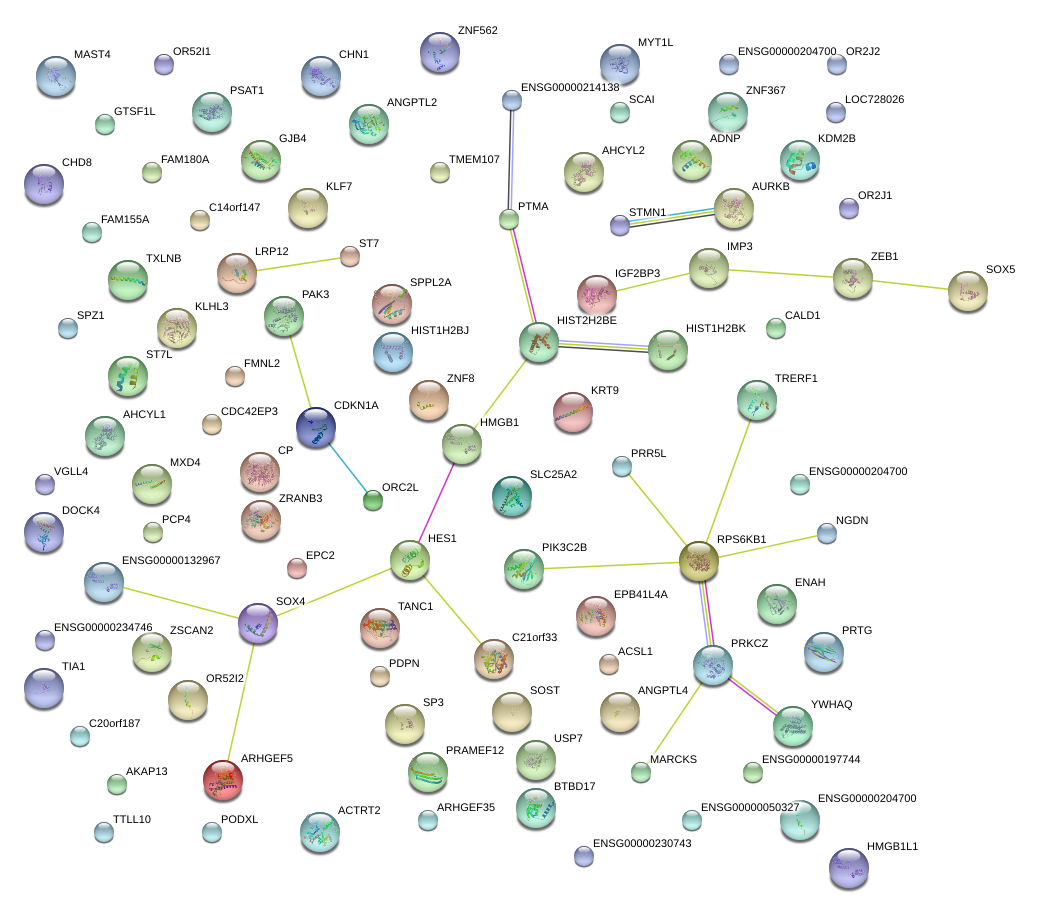
|
chr2_-_37752272
|
1.004
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr3_-_11736946
|
0.925
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr16_-_30841938
|
0.908
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr1_-_202730135
|
0.865
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr6_-_139654807
|
0.838
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr2_+_159533329
|
0.779
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr14_-_34001164
|
0.773
|
NM_138288
|
C14orf147
|
chromosome 14 open reading frame 147
|
|
chr7_+_128802719
|
0.766
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr12_-_24606616
|
0.749
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_+_232281470
|
0.716
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr6_+_114285209
|
0.699
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr15_-_73719546
|
0.673
|
NM_018285
|
IMP3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast)
|
|
chr2_+_152900076
|
0.664
|
|
FMNL2
|
formin-like 2
|
|
chr5_-_111782637
|
0.621
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr13_-_107317460
|
0.621
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr2_-_37752731
|
0.613
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr2_+_149118791
|
0.613
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr15_-_53822468
|
0.607
|
NM_173814
|
PRTG
|
protogenin
|
|
chr12_-_120389419
|
0.601
|
|
KDM2B
|
lysine (K)-specific demethylase 2B
|
|
chr2_-_175578156
|
0.589
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr13_-_29938029
|
0.587
|
NM_002128
|
HMGB1
|
high-mobility group box 1
|
|
chr2_+_232281491
|
0.575
|
|
PTMA
|
prothymosin, alpha
|
|
chr7_-_111633428
|
0.563
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr15_+_83724743
|
0.561
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr2_-_174538675
|
0.553
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr11_+_36274300
|
0.550
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr9_+_80101810
|
0.544
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr14_+_23008742
|
0.536
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr4_-_185963859
|
0.527
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr1_+_12757570
|
0.515
|
NM_001080830
|
PRAMEF12
|
PRAME family member 12
|
|
chr9_-_128910132
|
0.513
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr5_-_137099602
|
0.507
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr6_+_52637116
|
0.505
|
|
LOC730101
|
hypothetical LOC730101
|
|
chr1_+_34997928
|
0.498
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr17_-_36981830
|
0.497
|
NM_000226
|
KRT9
|
keratin 9
|
|
chr7_-_135083906
|
0.495
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr1_+_1104939
|
0.493
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr2_+_149119029
|
0.487
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr15_+_82945223
|
0.481
|
NM_001007072
NM_017894
NM_181877
|
ZSCAN2
|
zinc finger and SCAN domain containing 2
|
|
chr1_+_2927905
|
0.479
|
NM_080431
|
ACTRT2
|
actin-related protein T2
|
|
chr21_+_40161216
|
0.472
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chr14_+_23008723
|
0.472
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr6_+_21701942
|
0.470
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr9_-_126945536
|
0.460
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr4_-_2233606
|
0.458
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr6_-_27222556
|
0.458
|
NM_080593
|
HIST1H2BK
|
histone cluster 1, H2bk
|
|
chr17_-_69869552
|
0.454
|
NM_001080466
|
BTBD17
|
BTB (POZ) domain containing 17
|
|
chr1_+_13784553
|
0.452
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr6_+_29249289
|
0.448
|
NM_030905
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2
|
|
chr5_+_66160359
|
0.448
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr3_-_150422474
|
0.447
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr1_+_1994943
|
0.437
|
NM_001033581
|
PRKCZ
|
protein kinase C, zeta
|
|
chr2_-_136005238
|
0.436
|
NM_032143
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3
|
|
chr7_+_148613304
|
0.433
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr7_+_116447527
|
0.433
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr20_-_48980858
|
0.431
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr17_-_8054587
|
0.430
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr5_+_79651545
|
0.429
|
NM_032567
|
SPZ1
|
spermatogenic leucine zipper 1
|
|
chr2_-_234317362
|
0.428
|
NM_001001394
|
DNAJB3
|
DnaJ (Hsp40) homolog, subfamily B, member 3
|
|
chr4_-_2233504
|
0.428
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr11_+_4564596
|
0.428
|
NM_001005170
|
OR52I2
|
olfactory receptor, family 52, subfamily I, member 2
|
|
chr6_+_36752244
|
0.424
|
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr1_+_207668787
|
0.424
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr17_-_39191658
|
0.416
|
NM_025237
|
SOST
|
sclerostin
|
|
chr1_+_116455898
|
0.407
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr10_+_31650069
|
0.405
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr19_+_63482118
|
0.402
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr16_-_8964751
|
0.399
|
NM_003470
|
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated)
|
|
chr7_+_134114957
|
0.398
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_26105230
|
0.396
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr2_-_207739815
|
0.391
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr3_+_195336625
|
0.391
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr20_-_41788975
|
0.386
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr7_-_130891861
|
0.386
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr9_-_98220360
|
0.385
|
|
ZNF367
|
zinc finger protein 367
|
|
chr2_-_9688552
|
0.385
|
NM_006826
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr6_-_42527760
|
0.384
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr2_-_1905508
|
0.383
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr2_-_201536425
|
0.382
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr7_+_143683365
|
0.380
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr13_-_29937966
|
0.379
|
|
HMGB1
HMGB1P6
|
high-mobility group box 1
high-mobility group box 1 pseudogene 6
|
|
chr14_-_20975241
|
0.379
|
NM_020920
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr2_-_70329067
|
0.374
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr1_-_223907283
|
0.374
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr1_+_32777556
|
0.373
|
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr6_-_20320607
|
0.372
|
NM_001080480
|
MBOAT1
|
membrane bound O-acyltransferase domain containing 1
|
|
chr6_-_26767892
|
0.369
|
NM_024639
|
ZNF322A
|
zinc finger protein 322A
|
|
chrX_+_49013243
|
0.367
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr1_-_203557379
|
0.365
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr6_-_24985432
|
0.364
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr14_-_92284626
|
0.364
|
NM_001008530
NM_005606
|
LGMN
|
legumain
|
|
chr4_+_154345039
|
0.363
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr1_+_24158898
|
0.362
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr2_+_178893216
|
0.360
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_+_199975674
|
0.358
|
|
NAV1
|
neuron navigator 1
|
|
chr20_-_10602139
|
0.355
|
|
JAG1
|
jagged 1
|
|
chr20_+_42644633
|
0.354
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chrX_+_49013270
|
0.354
|
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr2_+_26862416
|
0.349
|
|
CENPA
|
centromere protein A
|
|
chr1_+_234372454
|
0.348
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr20_+_17628592
|
0.345
|
NM_001014977
NM_178477
|
BANF2
|
barrier to autointegration factor 2
|
|
chr1_+_43945804
|
0.344
|
NM_006279
NM_174963
NM_174964
NM_174965
NM_174966
NM_174967
NM_174968
NM_174969
NM_174970
NM_174971
|
ST3GAL3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3
|
|
chr17_-_70662088
|
0.343
|
|
HN1
|
hematological and neurological expressed 1
|
|
chr2_+_54536780
|
0.342
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr3_+_44571600
|
0.341
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr20_+_58004811
|
0.341
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chr18_-_13905534
|
0.338
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr9_+_124742932
|
0.337
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr2_+_85676347
|
0.333
|
NM_016494
|
RNF181
|
ring finger protein 181
|
|
chr1_+_183970093
|
0.333
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr6_-_29752909
|
0.332
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr3_+_100840129
|
0.332
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr15_-_32446537
|
0.329
|
NM_153613
|
LPCAT4
|
lysophosphatidylcholine acyltransferase 4
|
|
chr14_-_89155078
|
0.329
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr6_-_150388292
|
0.327
|
NM_130900
|
RAET1L
|
retinoic acid early transcript 1L
|
|
chr16_+_8675904
|
0.326
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr9_-_98220482
|
0.325
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr3_+_195336631
|
0.324
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr2_-_20413838
|
0.324
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr9_+_132990834
|
0.322
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr2_-_96769502
|
0.322
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr7_+_134114912
|
0.320
|
|
CALD1
|
caldesmon 1
|
|
chr22_-_37481857
|
0.319
|
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr5_+_148186348
|
0.318
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr17_-_36996611
|
0.316
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr2_+_62786290
|
0.314
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr10_+_170423
|
0.311
|
NM_001161482
NM_006624
NM_212479
|
ZMYND11
|
zinc finger, MYND-type containing 11
|
|
chr3_-_107070400
|
0.311
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr12_+_62524778
|
0.310
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr3_-_139315011
|
0.310
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr12_-_67012303
|
0.310
|
NM_017440
NM_020128
|
MDM1
|
Mdm1 nuclear protein homolog (mouse)
|
|
chr11_+_34029764
|
0.309
|
NM_005898
NM_203364
|
CAPRIN1
|
cell cycle associated protein 1
|
|
chr21_+_17807167
|
0.309
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr10_-_14686243
|
0.308
|
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr5_-_125956996
|
0.307
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr2_+_149895274
|
0.307
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr7_+_123083096
|
0.307
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr21_-_15358996
|
0.306
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr9_-_132759045
|
0.306
|
NM_198180
|
QRFP
|
pyroglutamylated RFamide peptide
|
|
chr9_+_132990760
|
0.306
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr3_-_194118627
|
0.304
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr2_-_171631728
|
0.304
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr5_+_108112537
|
0.303
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr16_-_8870343
|
0.303
|
NM_014316
|
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa
|
|
chr1_+_6767968
|
0.302
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr10_+_70418482
|
0.302
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr12_+_56374004
|
0.300
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr2_-_61618898
|
0.300
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr1_-_234096793
|
0.300
|
|
|
|
|
chr2_-_9688450
|
0.299
|
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide
|
|
chr7_+_116238309
|
0.299
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr19_-_17275083
|
0.298
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr17_-_45617899
|
0.297
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr5_+_14493905
|
0.294
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr12_-_102413875
|
0.292
|
NM_198521
NM_001099336
|
C12orf42
|
chromosome 12 open reading frame 42
|
|
chr6_-_53321545
|
0.292
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr4_+_87734489
|
0.292
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr7_-_27171600
|
0.289
|
|
HOXA9
|
homeobox A9
|
|
chr22_+_20316968
|
0.287
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr5_-_141318810
|
0.287
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr1_-_21373816
|
0.286
|
NM_001198803
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr15_-_42274529
|
0.285
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr3_+_113200696
|
0.284
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr20_+_54541672
|
0.284
|
NM_001013646
|
C20orf107
|
chromosome 20 open reading frame 107
|
|
chrX_-_76928334
|
0.283
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr2_+_85619802
|
0.282
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr6_-_152664906
|
0.282
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr3_+_123817151
|
0.282
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr16_-_10696285
|
0.280
|
NM_144674
|
TEKT5
|
tektin 5
|
|
chr15_+_75010997
|
0.280
|
NM_002902
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr1_-_6468108
|
0.280
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr6_+_31190505
|
0.279
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr16_-_78192111
|
0.279
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr6_+_27222839
|
0.278
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr2_+_206255376
|
0.277
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr11_+_6854431
|
0.276
|
NM_207186
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr17_-_46298671
|
0.276
|
|
TOB1
|
transducer of ERBB2, 1
|
|
chr2_+_152899837
|
0.276
|
NM_052905
|
FMNL2
|
formin-like 2
|
|
chr8_+_128497038
|
0.274
|
NM_001159542
|
POU5F1B
|
POU class 5 homeobox 1B
|
|
chr7_-_27171673
|
0.273
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr7_+_134114701
|
0.273
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr11_+_60448487
|
0.273
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr22_-_37481925
|
0.273
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr6_+_7671267
|
0.273
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr11_+_33520348
|
0.273
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr9_-_130980359
|
0.271
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr18_+_7936885
|
0.271
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr3_+_113200275
|
0.271
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr21_-_30460841
|
0.270
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr3_+_158637273
|
0.269
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr7_+_27102508
|
0.268
|
|
|
|
|
chr6_+_46728590
|
0.268
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr1_+_110683586
|
0.267
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr7_-_27171648
|
0.267
|
|
HOXA9
|
homeobox A9
|
|
chr16_-_20323533
|
0.267
|
NM_174924
|
PDILT
|
protein disulfide isomerase-like, testis expressed
|
|
chr2_+_149894744
|
0.266
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr9_+_102275310
|
0.265
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr2_-_73151894
|
0.264
|
|
SFXN5
|
sideroflexin 5
|
|
chr16_+_55373241
|
0.263
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr11_+_6823458
|
0.262
|
NM_178168
|
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5
|