|
chr11_-_16380967
|
3.708
|
NM_001145819
NM_017508
|
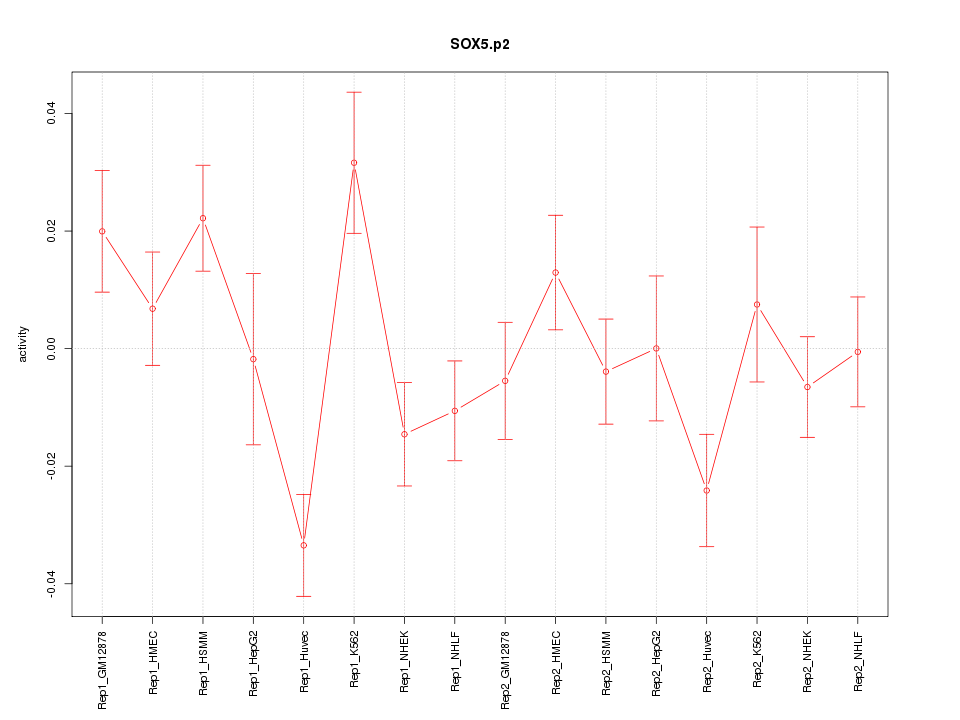
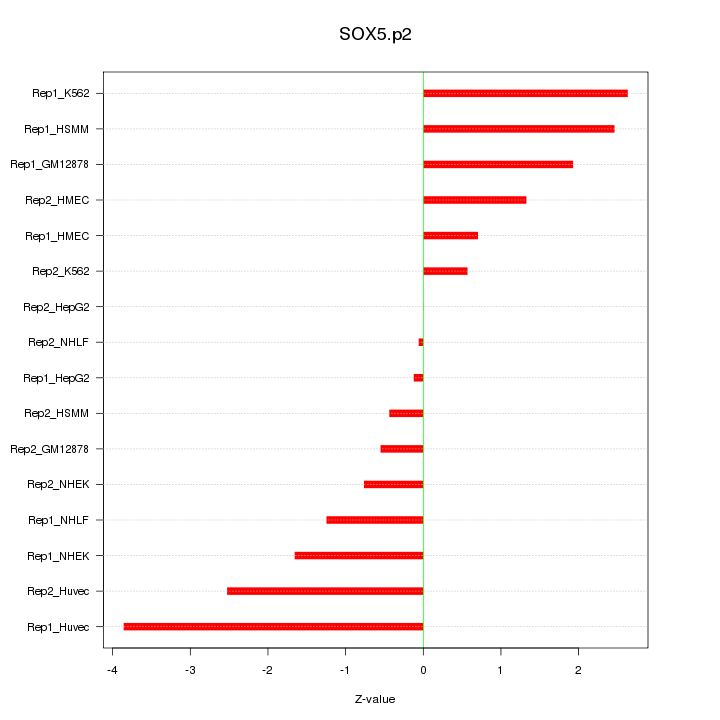
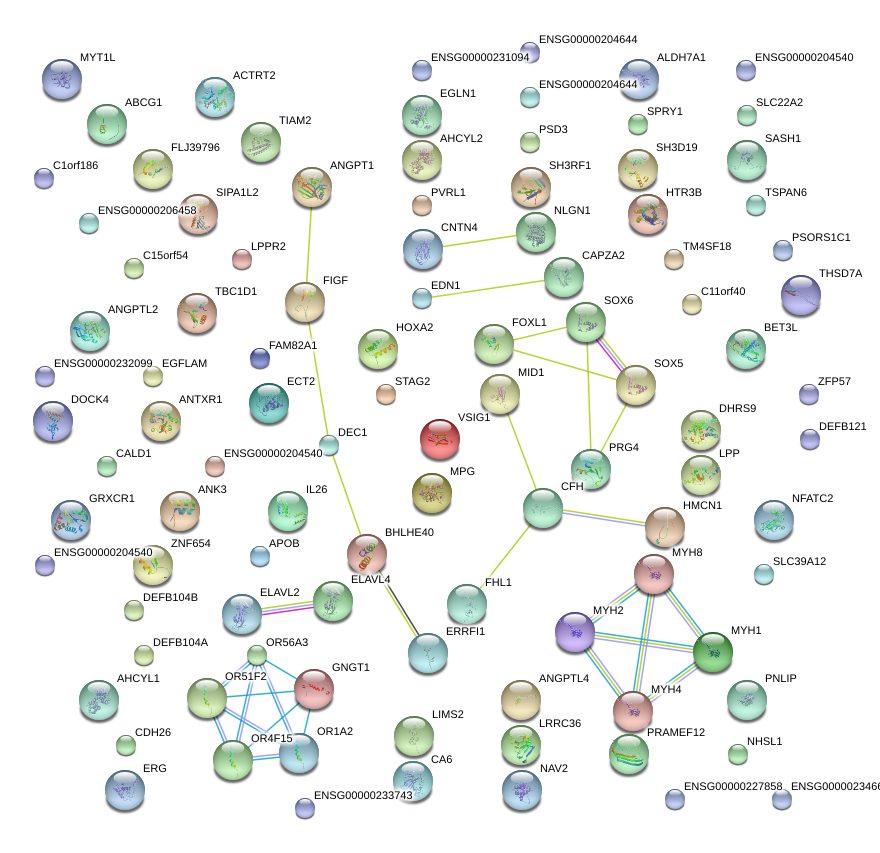
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr12_-_24606616
|
2.601
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr6_+_12398514
|
2.188
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr21_+_42492867
|
1.979
|
NM_207627
NM_207628
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1
|
|
chr6_+_21774624
|
1.949
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr3_+_174598937
|
1.921
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr7_+_128802719
|
1.781
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr11_+_20005704
|
1.776
|
|
NAV2
|
neuron navigator 2
|
|
chr1_+_50344550
|
1.743
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_-_111633428
|
1.738
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr7_-_11838260
|
1.676
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr11_-_119104463
|
1.618
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr1_+_183970093
|
1.565
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr5_-_125956996
|
1.534
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr17_-_10393664
|
1.345
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr16_+_65938753
|
1.331
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr21_-_38792173
|
1.329
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_66905802
|
1.313
|
NM_018402
|
IL26
|
interleukin 26
|
|
chr11_+_5925152
|
1.300
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr2_-_128138435
|
1.264
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr7_-_27108826
|
1.249
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr4_+_42590038
|
1.216
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr6_+_155579788
|
1.210
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr1_+_8928508
|
1.190
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chr7_+_134226690
|
1.177
|
NM_033139
NM_033140
|
CALD1
|
caldesmon 1
|
|
chr20_-_29457650
|
1.177
|
NM_001011878
|
DEFB121
|
defensin, beta 121
|
|
chr7_-_111633620
|
1.166
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr20_-_49612574
|
1.151
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr10_+_118295417
|
1.112
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chrX_-_99778340
|
1.109
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr1_+_12757570
|
1.106
|
NM_001080830
|
PRAMEF12
|
PRAME family member 12
|
|
chr1_-_7998265
|
1.095
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chrX_+_107174855
|
1.086
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr3_-_150533948
|
1.073
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chrX_-_10548458
|
1.069
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr20_-_49612745
|
1.066
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr3_+_189425886
|
1.063
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_+_38439399
|
1.061
|
NM_182798
NM_182799
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr11_+_113280727
|
1.032
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr2_-_21084077
|
1.031
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr10_-_61819493
|
1.030
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_2927905
|
1.020
|
NM_080431
|
ACTRT2
|
actin-related protein T2
|
|
chr3_+_173954940
|
1.020
|
NM_018098
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr2_+_38009405
|
1.009
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr6_+_31190505
|
0.998
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr1_-_229627412
|
0.997
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr7_+_134226727
|
0.994
|
|
CALD1
|
caldesmon 1
|
|
chr16_+_85169615
|
0.993
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr15_+_100175888
|
0.977
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr1_+_184532027
|
0.965
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr11_+_4799126
|
0.957
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr2_+_69093641
|
0.943
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr2_+_169631777
|
0.934
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr1_-_204455269
|
0.931
|
NM_001007544
|
C1orf186
|
chromosome 1 open reading frame 186
|
|
chr3_-_194118321
|
0.922
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr6_-_160599779
|
0.922
|
NM_003058
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2
|
|
chr6_-_138935359
|
0.908
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr6_+_148705421
|
0.906
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr6_+_148705817
|
0.895
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr6_-_116973465
|
0.895
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr5_-_168204430
|
0.895
|
|
|
|
|
chr2_-_1905508
|
0.885
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr8_-_18585721
|
0.865
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr10_+_18280773
|
0.864
|
NM_001145195
NM_152725
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chr5_-_71838875
|
0.862
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr9_-_128910132
|
0.853
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr11_-_4555625
|
0.845
|
NM_144663
|
C11orf40
|
chromosome 11 open reading frame 40
|
|
chrX_+_135079461
|
0.844
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr6_-_29752909
|
0.837
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr8_+_7731402
|
0.834
|
NM_080389
NM_001040702
|
DEFB104A
DEFB104B
|
defensin, beta 104A
defensin, beta 104B
|
|
chr4_-_152316105
|
0.829
|
NM_001128924
|
SH3D19
|
SH3 domain containing 19
|
|
chrX_-_15312390
|
0.827
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr3_+_88270951
|
0.822
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr7_+_93373755
|
0.815
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr1_+_559129
|
0.806
|
|
|
|
|
chr20_+_58004811
|
0.806
|
NM_021810
|
CDH26
|
cadherin 26
|
|
chr17_+_3047559
|
0.805
|
NM_012352
|
OR1A2
|
olfactory receptor, family 1, subfamily A, member 2
|
|
chrX_+_123062143
|
0.796
|
|
STAG2
|
stromal antigen 2
|
|
chr9_+_116943917
|
0.795
|
NM_017418
|
DEC1
|
deleted in esophageal cancer 1
|
|
chr1_-_230717850
|
0.794
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr15_+_37330176
|
0.783
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chr4_+_124537334
|
0.777
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_37655385
|
0.773
|
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr8_-_108579270
|
0.770
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr3_+_3056307
|
0.770
|
NM_175612
|
CNTN4
|
contactin 4
|
|
chr7_+_116238309
|
0.767
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr4_-_170314382
|
0.761
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr15_+_69220841
|
0.759
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr12_-_103846186
|
0.756
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr12_+_54172413
|
0.756
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr7_-_16811131
|
0.753
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr2_+_27573206
|
0.750
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr1_+_33494745
|
0.749
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chrX_-_50573759
|
0.730
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr3_-_194118627
|
0.730
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr6_-_138581319
|
0.729
|
NM_021635
|
PBOV1
|
prostate and breast cancer overexpressed 1
|
|
chr7_-_27171673
|
0.721
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr3_+_182900081
|
0.716
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr12_-_109842736
|
0.716
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr9_+_27099138
|
0.715
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr7_+_153633127
|
0.714
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr7_-_139948780
|
0.714
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chrX_+_130019896
|
0.713
|
NM_144967
|
ARHGAP36
|
Rho GTPase activating protein 36
|
|
chr1_-_154484341
|
0.708
|
NM_198406
NM_024897
|
PAQR6
|
progestin and adipoQ receptor family member VI
|
|
chr1_+_161305188
|
0.704
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr17_+_63252704
|
0.704
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr4_+_120029426
|
0.703
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr9_-_4142182
|
0.703
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_-_133144094
|
0.701
|
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr12_+_32546243
|
0.699
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr6_-_42527760
|
0.695
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr12_-_69289889
|
0.681
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr3_-_47909001
|
0.678
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr11_+_19328846
|
0.677
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr7_+_29486010
|
0.664
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr9_+_129066576
|
0.659
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr4_-_185376012
|
0.657
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr14_-_88953059
|
0.648
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr6_-_139654807
|
0.646
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr3_-_51976428
|
0.643
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr5_+_55182963
|
0.640
|
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr5_-_148738980
|
0.640
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr9_-_72673753
|
0.640
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr12_+_26096757
|
0.638
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr8_+_42671718
|
0.627
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr7_+_142850667
|
0.625
|
NM_177437
|
TAS2R60
|
taste receptor, type 2, member 60
|
|
chrX_-_19727828
|
0.615
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr13_+_23742846
|
0.615
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr10_-_116434364
|
0.611
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_151442542
|
0.604
|
NM_001010857
|
LELP1
|
late cornified envelope-like proline-rich 1
|
|
chrX_+_135079120
|
0.602
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_-_58627509
|
0.598
|
NM_138805
|
FAM3D
|
family with sequence similarity 3, member D
|
|
chr4_+_141397889
|
0.591
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr7_-_26198673
|
0.589
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chrX_+_15435351
|
0.588
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr15_+_57286313
|
0.588
|
NM_033195
|
LDHAL6B
|
lactate dehydrogenase A-like 6B
|
|
chr10_+_86121526
|
0.587
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr10_-_108914277
|
0.586
|
NM_001013031
NM_052918
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chrX_-_110541029
|
0.586
|
NM_000555
|
DCX
|
doublecortin
|
|
chr2_-_160181206
|
0.583
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr2_-_56266408
|
0.583
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr1_+_61697209
|
0.581
|
|
NFIA
|
nuclear factor I/A
|
|
chr7_+_111633878
|
0.580
|
NM_021994
|
ZNF277
|
zinc finger protein 277
|
|
chr18_-_27594696
|
0.580
|
NM_001034172
|
MCART2
|
mitochondrial carrier triple repeat 2
|
|
chr1_+_199975674
|
0.580
|
|
NAV1
|
neuron navigator 1
|
|
chr15_+_60146467
|
0.579
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chrX_+_107569671
|
0.575
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr1_-_202730135
|
0.575
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr6_-_29120930
|
0.573
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chr19_-_14800275
|
0.570
|
NM_017506
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5
|
|
chr9_-_129557373
|
0.570
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr6_-_132064233
|
0.569
|
NM_030908
|
OR2A4
|
olfactory receptor, family 2, subfamily A, member 4
|
|
chr1_+_61642287
|
0.569
|
|
NFIA
|
nuclear factor I/A
|
|
chr2_-_73170290
|
0.569
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr5_-_38631263
|
0.568
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr9_+_115265827
|
0.564
|
NM_017790
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr17_-_31332568
|
0.564
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr3_+_25444753
|
0.559
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_+_44571600
|
0.558
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr1_+_68070573
|
0.556
|
|
|
|
|
chr12_-_98072602
|
0.555
|
NM_020140
NM_181670
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr7_+_111633972
|
0.555
|
|
ZNF277
|
zinc finger protein 277
|
|
chr6_-_24985432
|
0.555
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr4_-_24523605
|
0.553
|
NM_001130726
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr1_-_85703198
|
0.551
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_-_160838397
|
0.550
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr11_+_5862076
|
0.549
|
NM_001005165
|
OR52E4
|
olfactory receptor, family 52, subfamily E, member 4
|
|
chr10_+_69539255
|
0.546
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr7_-_130891861
|
0.545
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr2_+_178893216
|
0.545
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr12_+_47498778
|
0.538
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr9_+_123501186
|
0.536
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr3_-_11585264
|
0.531
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr16_-_30705707
|
0.531
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr19_+_3958690
|
0.529
|
NM_015897
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr18_-_13905534
|
0.529
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr16_-_20271530
|
0.524
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr1_+_198975227
|
0.523
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|
|
chr19_-_56164628
|
0.522
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr6_-_24491498
|
0.521
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr1_-_68470585
|
0.520
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr4_-_184478920
|
0.520
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr6_-_138862271
|
0.518
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr12_+_54132264
|
0.516
|
NM_054105
|
OR6C2
|
olfactory receptor, family 6, subfamily C, member 2
|
|
chr7_-_143587653
|
0.516
|
NM_001005328
|
OR2A7
OR2A4
|
olfactory receptor, family 2, subfamily A, member 7
olfactory receptor, family 2, subfamily A, member 4
|
|
chr16_+_54072969
|
0.514
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr8_-_6783193
|
0.514
|
NM_001925
|
DEFA4
|
defensin, alpha 4, corticostatin
|
|
chr16_+_31105579
|
0.513
|
|
|
|
|
chr1_+_116727514
|
0.512
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr5_-_16989371
|
0.511
|
NM_012334
|
MYO10
|
myosin X
|
|
chr16_+_55373241
|
0.509
|
|
NUP93
|
nucleoporin 93kDa
|
|
chr9_-_96442377
|
0.508
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr3_-_126215069
|
0.505
|
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr13_-_93929719
|
0.505
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr6_-_18095672
|
0.500
|
NM_001105566
NM_001105567
NM_001105568
NM_022113
|
KIF13A
|
kinesin family member 13A
|
|
chr3_+_25444837
|
0.499
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr5_-_121842680
|
0.499
|
|
|
|
|
chr1_-_40140126
|
0.498
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr7_-_150555153
|
0.498
|
NM_005692
NM_007189
|
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2
|
|
chr1_-_224993498
|
0.493
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|