|
chr17_+_64922420
|
1.990
|
NM_002758
|
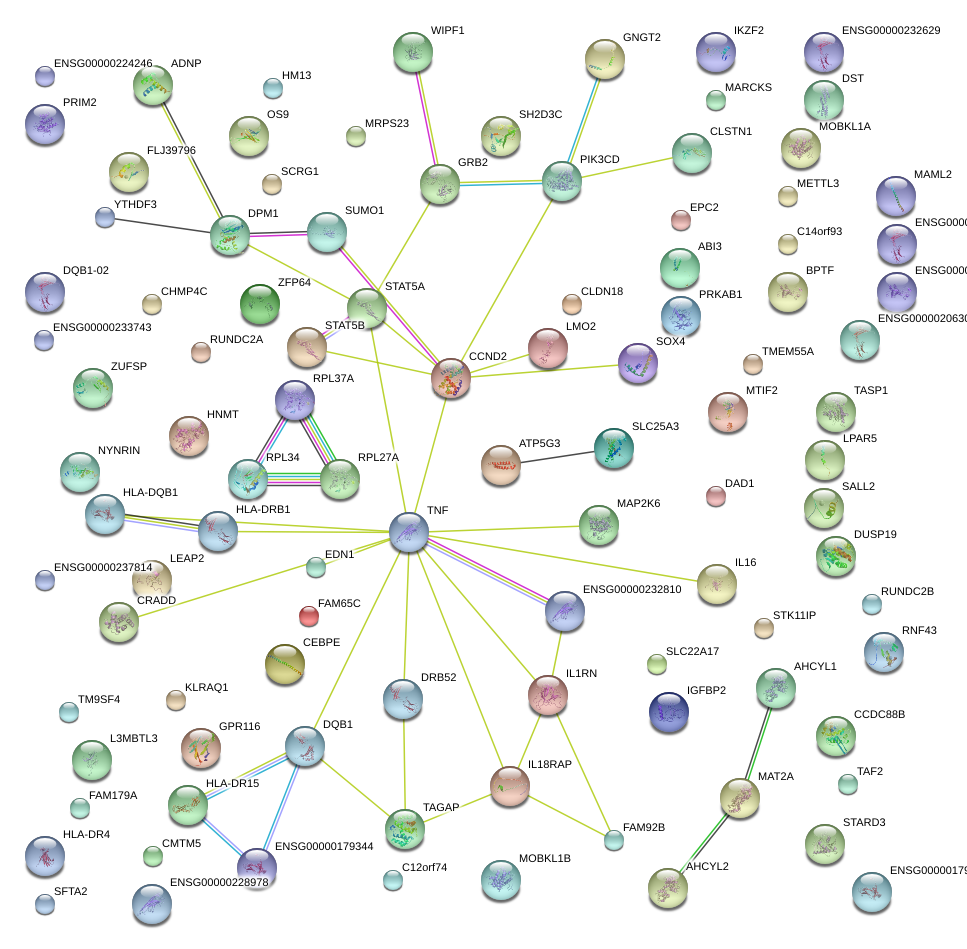
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr6_-_159386007
|
1.460
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr17_+_37693090
|
1.069
|
NM_003152
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr6_+_130381408
|
1.064
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr2_+_183651531
|
1.041
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr6_-_32742265
|
1.037
|
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr12_+_4253143
|
1.010
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr14_+_22915772
|
1.006
|
NM_001037288
NM_138460
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5
|
|
chr17_-_53847937
|
0.977
|
|
RNF43
|
ring finger protein 43
|
|
chr2_+_102401580
|
0.976
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr14_-_22658078
|
0.969
|
NM_001805
|
CEBPE
|
CCAAT/enhancer binding protein (C/EBP), epsilon
|
|
chr2_-_175207521
|
0.894
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr14_-_22127902
|
0.858
|
|
DAD1
|
defender against cell death 1
|
|
chr16_-_30841938
|
0.813
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr7_-_104440707
|
0.803
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr2_-_55349818
|
0.800
|
NM_001005369
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr17_+_35046843
|
0.784
|
NM_001165937
NM_001165938
NM_006804
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3
|
|
chr11_+_65026382
|
0.758
|
|
|
|
|
chr17_+_44643067
|
0.757
|
|
ABI3
|
ABI family, member 3
|
|
chr16_-_83703496
|
0.749
|
NM_198491
|
FAM92B
|
family with sequence similarity 92, member B
|
|
chr14_-_21074875
|
0.748
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr5_-_138822475
|
0.703
|
NM_001077693
|
ECSCR
|
endothelial cell-specific chemotaxis regulator
|
|
chr6_-_56816403
|
0.696
|
|
DST
|
dystonin
|
|
chr12_+_97513644
|
0.693
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr2_+_217071764
|
0.679
|
NM_000998
|
RPL37A
|
ribosomal protein L37a
|
|
chr2_+_138438277
|
0.674
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr12_-_120725210
|
0.671
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr2_-_55349851
|
0.658
|
NM_002453
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr17_-_44642934
|
0.656
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr6_+_12398514
|
0.644
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr2_+_220170804
|
0.640
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr4_+_109761170
|
0.638
|
NM_000995
NM_033625
|
RPL34
|
ribosomal protein L34
|
|
chr12_+_118590039
|
0.627
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr20_-_48686832
|
0.624
|
NM_080829
|
FAM65C
|
family with sequence similarity 65, member C
|
|
chr11_-_95715976
|
0.617
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr5_+_132237253
|
0.617
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr6_+_31651319
|
0.616
|
NM_000594
|
TNF
|
tumor necrosis factor
|
|
chr20_-_48980858
|
0.609
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr17_-_70901303
|
0.608
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr5_-_71838875
|
0.602
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr2_-_213723298
|
0.597
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr6_+_114285209
|
0.581
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr6_-_47030492
|
0.575
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr17_+_55269974
|
0.573
|
|
|
|
|
chr2_+_48521411
|
0.571
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr20_-_50241581
|
0.568
|
|
ZFP64
|
zinc finger protein 64 homolog (mouse)
|
|
chr16_+_28413421
|
0.565
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr17_-_53282376
|
0.564
|
NM_016070
|
MRPS23
|
mitochondrial ribosomal protein S23
|
|
chr2_+_149118791
|
0.560
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_+_217206450
|
0.542
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr14_-_22891865
|
0.540
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr6_+_57290357
|
0.535
|
NM_000947
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa)
|
|
chr8_-_92122219
|
0.534
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr20_-_13567529
|
0.533
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr15_+_79376295
|
0.527
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr14_-_22127954
|
0.525
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr14_+_23937766
|
0.524
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr16_+_11978002
|
0.520
|
NM_032167
|
RUNDC2A
|
RUN domain containing 2A
|
|
chr2_+_149119029
|
0.519
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr7_+_128651976
|
0.514
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr2_-_175754366
|
0.511
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr8_-_120914233
|
0.509
|
NM_003184
|
TAF2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa
|
|
chr6_+_21701942
|
0.503
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr14_-_21049204
|
0.499
|
NM_019852
|
METTL3
|
methyltransferase like 3
|
|
chr6_-_117096594
|
0.497
|
NM_145062
|
ZUFSP
|
zinc finger with UFM1-specific peptidase domain
|
|
chr4_-_174557191
|
0.493
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr11_+_8660913
|
0.492
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr3_+_139200347
|
0.491
|
NM_001002026
|
CLDN18
|
claudin 18
|
|
chr6_-_31007905
|
0.490
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr14_-_22549168
|
0.489
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr9_-_129580732
|
0.485
|
NM_170600
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr2_+_113601608
|
0.480
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr20_+_29598848
|
0.478
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr12_+_91620749
|
0.476
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr12_-_6615348
|
0.474
|
NM_020400
|
LPAR5
|
lysophosphatidic acid receptor 5
|
|
chr2_+_113601616
|
0.473
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr8_+_82807218
|
0.472
|
NM_152284
|
CHMP4C
|
chromatin modifying protein 4C
|
|
chr12_+_92595281
|
0.471
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr1_+_9634368
|
0.471
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr20_+_30160952
|
0.468
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr11_+_63864263
|
0.466
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr12_+_56374004
|
0.466
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr8_-_92151363
|
0.462
|
|
|
|
|
chr2_+_85619802
|
0.458
|
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr2_+_29057667
|
0.457
|
NM_199280
|
FAM179A
|
family with sequence similarity 179, member A
|
|
chr20_-_49008445
|
0.447
|
NM_003859
|
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit
|
|
chr2_-_202811459
|
0.445
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chr11_-_33847507
|
0.442
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr17_+_63252704
|
0.440
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr2_-_74259308
|
0.438
|
NM_018221
|
MOBKL1B
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr8_+_64243741
|
0.433
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr14_-_21172837
|
0.432
|
NM_001005466
|
OR10G2
|
olfactory receptor, family 10, subfamily G, member 2
|
|
chr6_+_31662454
|
0.431
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr7_+_104959767
|
0.431
|
NM_021930
|
RINT1
|
RAD50 interactor 1
|
|
chr16_-_29781842
|
0.425
|
NM_006319
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
|
|
chr11_+_101693403
|
0.423
|
NM_001165
NM_182962
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr17_-_41625921
|
0.423
|
NM_001193466
|
KIAA1267
|
KIAA1267
|
|
chr12_-_15005761
|
0.420
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr6_-_56824371
|
0.417
|
|
DST
|
dystonin
|
|
chr7_+_141084591
|
0.414
|
NM_003143
|
SSBP1
|
single-stranded DNA binding protein 1
|
|
chr14_+_28311740
|
0.413
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr19_-_47623340
|
0.413
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr8_-_57149588
|
0.413
|
NM_001023
NM_001146227
|
RPS20
|
ribosomal protein S20
|
|
chr6_+_31661934
|
0.411
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr16_-_15644465
|
0.411
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr5_+_61910318
|
0.411
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr16_-_3246587
|
0.410
|
NM_000243
NM_001198536
|
MEFV
|
Mediterranean fever
|
|
chr16_+_15644624
|
0.407
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr16_+_24696069
|
0.407
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr1_+_159462456
|
0.406
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr5_-_158567411
|
0.406
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr12_-_56157898
|
0.404
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr14_-_20969706
|
0.403
|
NM_001170629
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr6_-_116488458
|
0.402
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr1_+_154390939
|
0.401
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_-_205210567
|
0.401
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr7_+_139175420
|
0.400
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr4_-_103968104
|
0.400
|
NM_003340
NM_181886
NM_181888
NM_181889
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
|
|
chr20_+_30161173
|
0.399
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr2_-_27147995
|
0.399
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr2_-_206659140
|
0.399
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr2_-_190893015
|
0.397
|
NM_014362
NM_198047
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase
|
|
chr11_+_74788200
|
0.395
|
NM_001005
|
RPS3
|
ribosomal protein S3
|
|
chr6_-_119137923
|
0.392
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr16_-_87460514
|
0.391
|
NM_001080487
|
PABPN1L
|
poly(A) binding protein, nuclear 1-like (cytoplasmic)
|
|
chr1_-_171441234
|
0.390
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr20_-_50241930
|
0.387
|
NM_018197
NM_022088
NM_199426
NM_199427
|
ZFP64
|
zinc finger protein 64 homolog (mouse)
|
|
chr11_+_65027721
|
0.387
|
|
|
|
|
chr2_+_183651713
|
0.382
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr6_+_15354278
|
0.382
|
NM_004973
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr7_+_102775309
|
0.381
|
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr2_-_178125680
|
0.380
|
NM_152517
|
TTC30B
|
tetratricopeptide repeat domain 30B
|
|
chr6_-_34963565
|
0.378
|
NM_005643
|
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa
|
|
chr1_-_220829704
|
0.378
|
NM_005681
NM_139352
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa
|
|
chr2_-_96174865
|
0.374
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr6_-_28327980
|
0.374
|
NM_019110
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4
|
|
chr22_+_29333069
|
0.373
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr14_-_24047261
|
0.370
|
NM_001836
|
CMA1
|
chymase 1, mast cell
|
|
chr20_+_47096280
|
0.368
|
|
CSE1L
|
CSE1 chromosome segregation 1-like (yeast)
|
|
chr14_-_20975241
|
0.366
|
NM_020920
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr18_+_30875297
|
0.366
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr5_-_139402909
|
0.365
|
|
NRG2
|
neuregulin 2
|
|
chr5_+_180514548
|
0.365
|
NM_206880
|
OR2V2
|
olfactory receptor, family 2, subfamily V, member 2
|
|
chr5_+_139485703
|
0.363
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr7_+_99052489
|
0.362
|
NM_145115
|
ZNF498
|
zinc finger protein 498
|
|
chr6_-_32265489
|
0.362
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr2_-_61551403
|
0.359
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr12_-_46438447
|
0.358
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr17_+_44642587
|
0.358
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr5_+_143171918
|
0.357
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr17_+_35718948
|
0.357
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr22_+_35586950
|
0.357
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr6_+_43136159
|
0.355
|
NM_201522
|
KLC4
|
kinesin light chain 4
|
|
chr6_+_31647854
|
0.353
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr16_+_24459173
|
0.351
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_-_116184846
|
0.349
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr3_+_87359088
|
0.349
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr9_-_6005602
|
0.349
|
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr17_+_35097851
|
0.349
|
NM_001005862
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr2_+_85619797
|
0.349
|
NM_005911
|
MAT2A
|
methionine adenosyltransferase II, alpha
|
|
chr16_+_51690564
|
0.349
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr12_-_56111054
|
0.347
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr2_-_242449662
|
0.345
|
NM_005018
|
PDCD1
|
programmed cell death 1
|
|
chr14_+_22410795
|
0.345
|
NM_014045
|
LRP10
|
low density lipoprotein receptor-related protein 10
|
|
chr2_-_24995402
|
0.344
|
NM_004036
|
ADCY3
|
adenylate cyclase 3
|
|
chr1_+_182040837
|
0.344
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr6_+_31731647
|
0.343
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr17_+_54642152
|
0.341
|
NM_018149
|
C17orf71
|
chromosome 17 open reading frame 71
|
|
chr12_+_52132353
|
0.340
|
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr17_-_44058612
|
0.339
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr2_-_174537021
|
0.338
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr2_-_180580024
|
0.337
|
NM_020943
|
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr4_+_39376058
|
0.336
|
NM_001111112
NM_001111113
NM_005339
|
UBE2K
|
ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast)
|
|
chr14_+_38714111
|
0.336
|
NM_002687
|
PNN
|
pinin, desmosome associated protein
|
|
chr12_+_52689062
|
0.335
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr14_+_22859236
|
0.334
|
NM_004643
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr13_-_47885018
|
0.334
|
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr22_-_35494019
|
0.333
|
NM_001177702
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr6_-_31658044
|
0.332
|
NM_002341
NM_009588
|
LTB
|
lymphotoxin beta (TNF superfamily, member 3)
|
|
chr5_+_133870175
|
0.332
|
|
|
|
|
chr4_+_84596141
|
0.332
|
NM_016067
|
MRPS18C
|
mitochondrial ribosomal protein S18C
|
|
chr10_+_22580992
|
0.331
|
|
|
|
|
chr6_-_25044156
|
0.330
|
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr3_+_98966253
|
0.328
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr17_+_24742068
|
0.326
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr2_+_112956204
|
0.325
|
NM_153712
|
TTL
|
tubulin tyrosine ligase
|
|
chr11_-_14342627
|
0.325
|
NM_001177314
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr17_-_59403932
|
0.325
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr7_-_30032649
|
0.325
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr10_+_104580420
|
0.320
|
|
|
|
|
chr2_-_190892730
|
0.320
|
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase
|
|
chr6_-_33016561
|
0.320
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr10_+_129735796
|
0.319
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr2_-_219404755
|
0.319
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr17_-_7061641
|
0.319
|
NM_001128827
|
DLG4
|
discs, large homolog 4 (Drosophila)
|
|
chr1_+_24518501
|
0.317
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_-_199348316
|
0.315
|
NM_000069
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr2_-_216944911
|
0.313
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr1_+_204797516
|
0.313
|
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr11_+_76423082
|
0.312
|
NM_138706
|
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase)
|