|
chr12_-_73889481
|
0.685
|
|
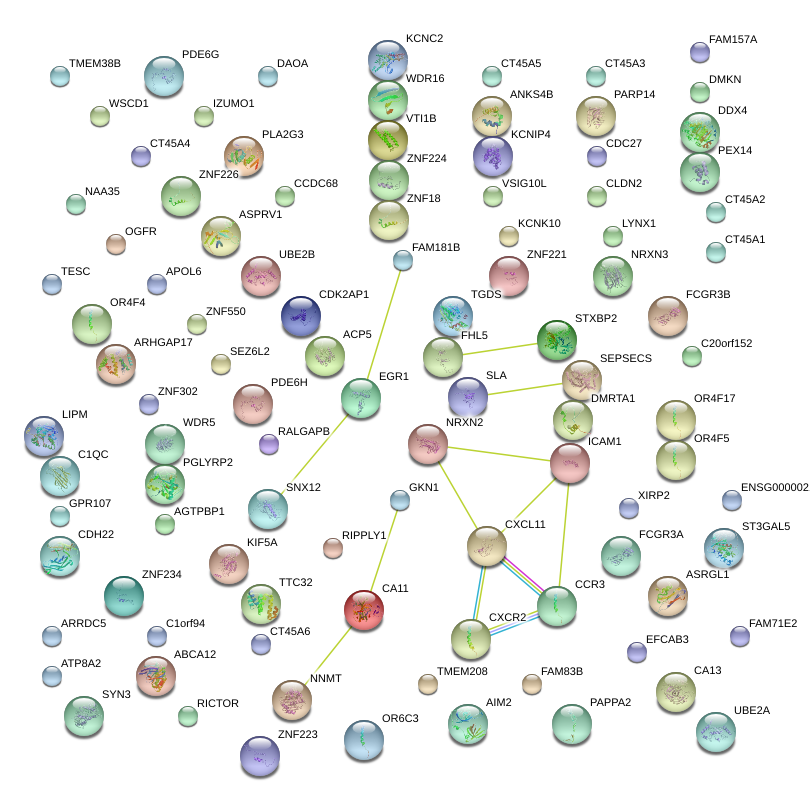
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr20_+_34019942
|
0.596
|
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr19_+_10242763
|
0.556
|
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr22_-_29866339
|
0.481
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr11_+_61861349
|
0.367
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr2_+_218698242
|
0.354
|
NM_001168298
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr4_-_77176163
|
0.347
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr9_+_107496638
|
0.333
|
NM_018112
|
TMEM38B
|
transmembrane protein 38B
|
|
chr18_-_50777634
|
0.330
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr2_-_70042900
|
0.316
|
NM_152792
|
ASPRV1
|
aspartic peptidase, retroviral-like 1
|
|
chr8_-_143856640
|
0.295
|
NM_023946
NM_177457
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr11_+_61861458
|
0.282
|
|
ASRGL1
|
asparaginase like 1
|
|
chr19_-_4853878
|
0.281
|
NM_001080523
|
ARRDC5
|
arrestin domain containing 5
|
|
chr13_+_24844084
|
0.268
|
NM_016529
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr2_-_215605054
|
0.262
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr19_-_60566419
|
0.255
|
NM_001145402
|
FAM71E2
|
family with sequence similarity 71, member E2
|
|
chr9_+_135994734
|
0.253
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr11_-_64247230
|
0.251
|
NM_015080
NM_138732
|
NRXN2
|
neurexin 2
|
|
chr19_+_49309350
|
0.247
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr6_+_54819527
|
0.237
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr3_+_46258875
|
0.228
|
NM_001164680
NM_001837
NM_178328
NM_178329
|
CCR3
|
chemokine (C-C motif) receptor 3
|
|
chrX_-_106033202
|
0.228
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr20_-_44370537
|
0.226
|
|
CDH22
|
cadherin 22, type 2
|
|
chr1_+_174698929
|
0.222
|
NM_020318
NM_021936
|
PAPPA2
|
pappalysin 2
|
|
chr1_+_22842704
|
0.220
|
NM_001114101
NM_172369
|
C1QC
|
complement component 1, q subcomponent, C chain
|
|
chr8_+_86344984
|
0.220
|
|
CA13
|
carbonic anhydrase XIII
|
|
chr5_+_55098469
|
0.218
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr8_+_86344945
|
0.211
|
NM_198584
|
CA13
|
carbonic anhydrase XIII
|
|
chr1_-_159786150
|
0.208
|
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr19_-_11550576
|
0.200
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr9_-_87546620
|
0.200
|
NM_015239
|
AGTPBP1
|
ATP/GTP binding protein 1
|
|
chr9_+_131937163
|
0.193
|
|
GPR107
|
G protein-coupled receptor 107
|
|
chr20_+_60906605
|
0.190
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr1_+_34405070
|
0.182
|
NM_032884
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr17_+_57811645
|
0.175
|
NM_173503
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr19_+_39860398
|
0.171
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr12_-_116021473
|
0.170
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr16_-_24876357
|
0.167
|
|
ARHGAP17
|
Rho GTPase activating protein 17
|
|
chr17_+_9420668
|
0.165
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chr14_-_67211081
|
0.165
|
NM_006370
|
VTI1B
|
vesicle transport through interaction with t-SNAREs homolog 1B (yeast)
|
|
chr8_-_143856179
|
0.163
|
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr17_+_5913149
|
0.162
|
|
WSCD1
|
WSC domain containing 1
|
|
chr13_+_104916216
|
0.162
|
NM_001161814
NM_172370
|
DAOA
|
D-amino acid oxidase activator
|
|
chr1_-_157313179
|
0.160
|
NM_004833
|
AIM2
|
absent in melanoma 2
|
|
chr19_-_56537189
|
0.159
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr19_-_53841064
|
0.158
|
NM_001217
|
CA11
|
carbonic anhydrase XI
|
|
chr2_-_19965095
|
0.152
|
NM_001008237
|
TTC32
|
tetratricopeptide repeat domain 32
|
|
chr2_-_85948287
|
0.149
|
NM_001042437
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr16_-_29817841
|
0.147
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr22_-_31732648
|
0.143
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr14_-_87862676
|
0.142
|
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr22_+_34374358
|
0.138
|
NM_030641
|
APOL6
|
apolipoprotein L, 6
|
|
chrX_+_106030049
|
0.135
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr8_+_86345256
|
0.135
|
|
CA13
|
carbonic anhydrase XIII
|
|
chr13_-_94046483
|
0.133
|
NM_014305
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr11_+_113672407
|
0.133
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr19_+_10242516
|
0.133
|
NM_000201
|
ICAM1
|
intercellular adhesion molecule 1
|
|
chr10_+_90552466
|
0.133
|
NM_001128215
|
LIPM
|
lipase, family member M
|
|
chr12_+_15017222
|
0.131
|
NM_006205
|
PDE6H
|
phosphodiesterase 6H, cGMP-specific, cone, gamma
|
|
chr19_+_49290319
|
0.130
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr6_+_97117144
|
0.130
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr19_+_49337548
|
0.129
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr22_+_34374421
|
0.127
|
|
APOL6
|
apolipoprotein L, 6
|
|
chr11_+_113672378
|
0.124
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr2_+_167468220
|
0.123
|
NM_001079810
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr4_-_21155376
|
0.122
|
NM_001035004
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr19_-_15451203
|
0.122
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr19_-_49309102
|
0.120
|
|
LOC100379224
|
hypothetical LOC100379224
|
|
chr1_-_159786441
|
0.119
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr9_+_87745876
|
0.116
|
NM_024635
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr16_+_88696172
|
0.114
|
|
FAM157C
|
family with sequence similarity 157, member C
|
|
chr9_+_22437353
|
0.114
|
|
DMRTA1
|
DMRT-like family A1
|
|
chr5_+_133734768
|
0.110
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr13_+_104916565
|
0.109
|
NM_001161812
|
DAOA
|
D-amino acid oxidase activator
|
|
chr3_+_123882355
|
0.108
|
NM_017554
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14
|
|
chr12_-_122322117
|
0.105
|
NM_004642
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1
|
|
chr5_+_133735004
|
0.104
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr13_-_94046402
|
0.104
|
|
TGDS
|
TDP-glucose 4,6-dehydratase
|
|
chr1_+_10457579
|
0.103
|
NM_004565
|
PEX14
|
peroxisomal biogenesis factor 14
|
|
chr19_-_62759536
|
0.101
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr19_+_49248003
|
0.098
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr8_+_86345274
|
0.097
|
|
|
|
|
chr16_+_65818511
|
0.094
|
NM_014187
|
TMEM208
|
transmembrane protein 208
|
|
chr1_+_58899
|
0.094
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chrX_+_134693879
|
0.093
|
NM_001017436
NM_152582
|
CT45A3
CT45A4
CT45A2
|
cancer/testis antigen family 45, member A3
cancer/testis antigen family 45, member A4
cancer/testis antigen family 45, member A2
|
|
chr9_+_87745922
|
0.092
|
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr19_-_53941835
|
0.092
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr12_+_54011750
|
0.089
|
NM_054104
|
OR6C3
|
olfactory receptor, family 6, subfamily C, member 3
|
|
chr8_-_134184491
|
0.088
|
NM_001045556
NM_001045557
|
SLA
|
Src-like-adaptor
|
|
chr17_-_11841413
|
0.087
|
NM_144680
|
ZNF18
|
zinc finger protein 18
|
|
chr5_-_39110251
|
0.087
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr19_-_40693176
|
0.086
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr20_+_36534871
|
0.085
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr20_-_44313731
|
0.081
|
NM_021248
|
CDH22
|
cadherin 22, type 2
|
|
chr1_-_205186330
|
0.079
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr9_-_85761451
|
0.076
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr1_+_198278339
|
0.075
|
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr3_+_130081130
|
0.074
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr19_-_5790702
|
0.072
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr9_-_23811477
|
0.072
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_11841476
|
0.072
|
NM_002521
|
NPPB
|
natriuretic peptide B
|
|
chr1_+_205329206
|
0.071
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr2_+_17798860
|
0.070
|
NM_001130009
|
LOC284952
GEN1
|
hypothetical protein LOC284952
Gen homolog 1, endonuclease (Drosophila)
|
|
chr19_+_46992361
|
0.070
|
NM_001815
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3
|
|
chr10_+_91142301
|
0.070
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr2_-_200424140
|
0.069
|
|
LOC348751
|
hypothetical LOC348751
|
|
chr9_-_85761472
|
0.069
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr1_+_205328834
|
0.067
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr12_-_74711642
|
0.067
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr19_+_49337568
|
0.066
|
|
ZNF234
|
zinc finger protein 234
|
|
chr5_-_133734616
|
0.065
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr5_-_39110235
|
0.062
|
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr11_-_62218759
|
0.062
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr19_-_4782711
|
0.061
|
NM_182919
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chr10_+_70699761
|
0.060
|
NM_033497
NM_033498
NM_033500
|
HK1
|
hexokinase 1
|
|
chr14_-_93988838
|
0.059
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr11_-_7684516
|
0.058
|
NM_198185
|
OVCH2
|
ovochymase 2 (gene/pseudogene)
|
|
chr3_+_130081149
|
0.058
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr4_-_145281237
|
0.057
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr9_+_22437064
|
0.057
|
|
|
|
|
chr3_+_130081161
|
0.057
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr1_-_159867781
|
0.057
|
NM_000570
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr12_-_7153068
|
0.056
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr2_-_113310796
|
0.055
|
NM_000576
|
IL1B
|
interleukin 1, beta
|
|
chr1_-_173428455
|
0.055
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr9_-_35608300
|
0.054
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr5_-_139993469
|
0.054
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr15_+_56217686
|
0.053
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr9_+_129226535
|
0.053
|
|
ZNF79
|
zinc finger protein 79
|
|
chr11_-_121492010
|
0.053
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr5_+_175807992
|
0.053
|
|
FAF2
|
Fas associated factor family member 2
|
|
chr3_+_133798770
|
0.053
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chrX_-_154216929
|
0.053
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr1_+_240078655
|
0.052
|
NM_003686
NM_006027
|
EXO1
|
exonuclease 1
|
|
chr19_-_11311254
|
0.052
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr2_+_170074457
|
0.052
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr5_+_175807986
|
0.052
|
|
FAF2
|
Fas associated factor family member 2
|
|
chr19_+_49268106
|
0.051
|
NM_001037813
|
ZNF284
|
zinc finger protein 284
|
|
chr3_+_130081210
|
0.051
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr5_+_175807978
|
0.050
|
|
FAF2
|
Fas associated factor family member 2
|
|
chr19_+_49221333
|
0.049
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr15_-_40173949
|
0.049
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr5_+_134237325
|
0.048
|
NM_024715
|
TXNDC15
|
thioredoxin domain containing 15
|
|
chr6_-_31007905
|
0.048
|
NM_205854
|
SFTA2
|
surfactant associated 2
|
|
chr3_-_147805715
|
0.047
|
|
PLSCR5
|
phospholipid scramblase family, member 5
|
|
chr20_+_35406497
|
0.047
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr3_-_42719322
|
0.047
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chr17_-_45901173
|
0.044
|
NM_001267
|
CHAD
|
chondroadherin
|
|
chr2_+_102401580
|
0.044
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr3_+_116824840
|
0.044
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr20_+_24397834
|
0.044
|
NM_024893
|
TMEM90B
|
transmembrane protein 90B
|
|
chr1_-_151788341
|
0.043
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr19_+_61798434
|
0.043
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr4_-_129429433
|
0.043
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr17_+_29707583
|
0.042
|
NM_005408
|
CCL13
|
chemokine (C-C motif) ligand 13
|
|
chr17_+_18796202
|
0.042
|
NM_001042450
NM_152351
|
SLC5A10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10
|
|
chr1_+_154389945
|
0.041
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr10_-_62882930
|
0.041
|
|
TMEM26
|
transmembrane protein 26
|
|
chr9_-_136949531
|
0.040
|
NM_002003
|
FCN1
|
ficolin (collagen/fibrinogen domain containing) 1
|
|
chr1_-_71285847
|
0.040
|
NM_001126044
NM_198714
NM_198715
NM_198716
NM_198717
NM_198718
NM_198719
|
PTGER3
|
prostaglandin E receptor 3 (subtype EP3)
|
|
chr19_+_49408523
|
0.039
|
NM_182490
|
ZNF227
|
zinc finger protein 227
|
|
chr3_+_134978615
|
0.038
|
|
TF
|
transferrin
|
|
chr17_+_29670171
|
0.038
|
NM_005623
|
CCL8
|
chemokine (C-C motif) ligand 8
|
|
chr3_+_23962518
|
0.037
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_-_45813116
|
0.037
|
|
SLC6A20
|
solute carrier family 6 (proline IMINO transporter), member 20
|
|
chr19_-_49644470
|
0.037
|
NM_014518
|
ZNF229
|
zinc finger protein 229
|
|
chr19_+_40431122
|
0.036
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr3_+_130081187
|
0.036
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr13_+_38159172
|
0.036
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr1_-_156056456
|
0.036
|
NM_001159397
NM_001159398
NM_052938
|
FCRL1
|
Fc receptor-like 1
|
|
chr3_+_12305336
|
0.036
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr5_+_175807961
|
0.036
|
NM_014613
|
FAF2
|
Fas associated factor family member 2
|
|
chr3_+_48456667
|
0.036
|
NM_015933
|
CCDC72
|
coiled-coil domain containing 72
|
|
chr12_-_8271460
|
0.035
|
NM_018088
|
FAM90A1
|
family with sequence similarity 90, member A1
|
|
chr5_+_31834787
|
0.034
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr17_+_71586857
|
0.034
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr12_-_108399510
|
0.033
|
NM_031954
|
KCTD10
|
potassium channel tetramerisation domain containing 10
|
|
chr10_+_85994788
|
0.033
|
NM_001012720
NM_001012722
NM_002921
|
RGR
|
retinal G protein coupled receptor
|
|
chr22_-_40856821
|
0.032
|
NM_000106
NM_001025161
|
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6
|
|
chr2_-_119998271
|
0.032
|
|
SCTR
|
secretin receptor
|
|
chr12_+_22669352
|
0.032
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chrX_+_128942435
|
0.032
|
|
|
|
|
chr16_-_65818221
|
0.031
|
NM_012163
|
LRRC29
|
leucine rich repeat containing 29
|
|
chr5_-_111340421
|
0.031
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr17_-_564698
|
0.031
|
NM_001128159
NM_018289
|
VPS53
|
vacuolar protein sorting 53 homolog (S. cerevisiae)
|
|
chr8_-_58068697
|
0.031
|
|
IMPAD1
|
inositol monophosphatase domain containing 1
|
|
chr5_+_139924224
|
0.031
|
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr8_-_42742729
|
0.031
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr4_+_25267183
|
0.031
|
NM_001177999
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr4_+_41235077
|
0.031
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_-_161380562
|
0.031
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr16_+_4614830
|
0.031
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr10_+_93548162
|
0.031
|
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2
|
|
chr10_+_95838801
|
0.030
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr12_+_2938703
|
0.030
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr17_-_70480385
|
0.030
|
NM_030630
|
C17orf28
|
chromosome 17 open reading frame 28
|
|
chr8_-_104496549
|
0.030
|
NM_030780
|
SLC25A32
|
solute carrier family 25, member 32
|
|
chr22_-_29833484
|
0.030
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr1_+_35020425
|
0.030
|
NM_001005752
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr14_+_102313585
|
0.029
|
|
TRAF3
|
TNF receptor-associated factor 3
|