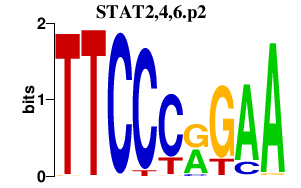
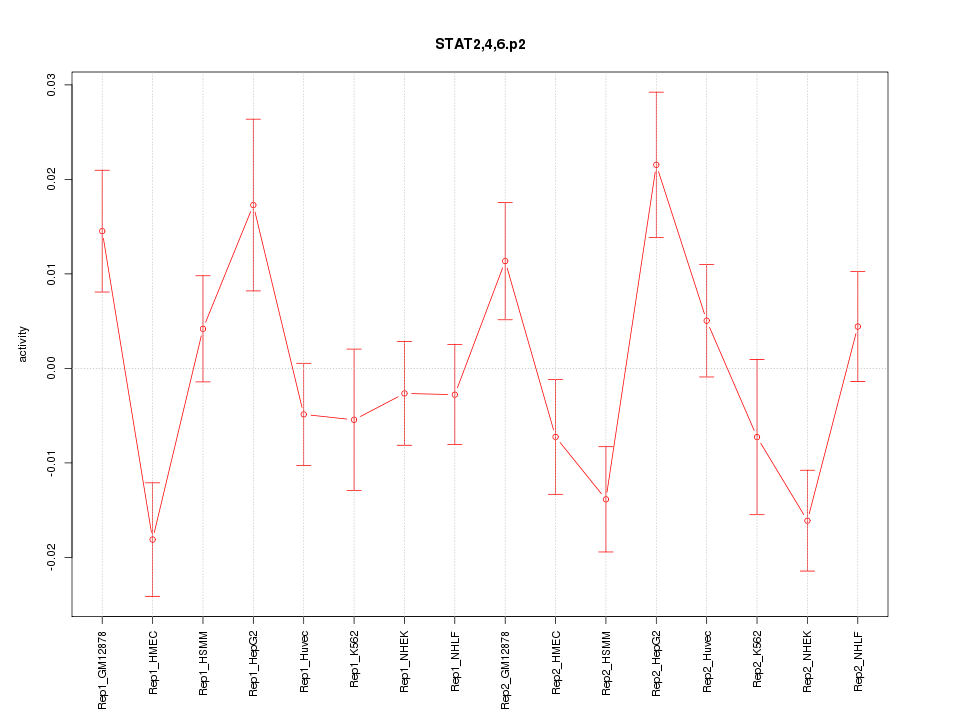
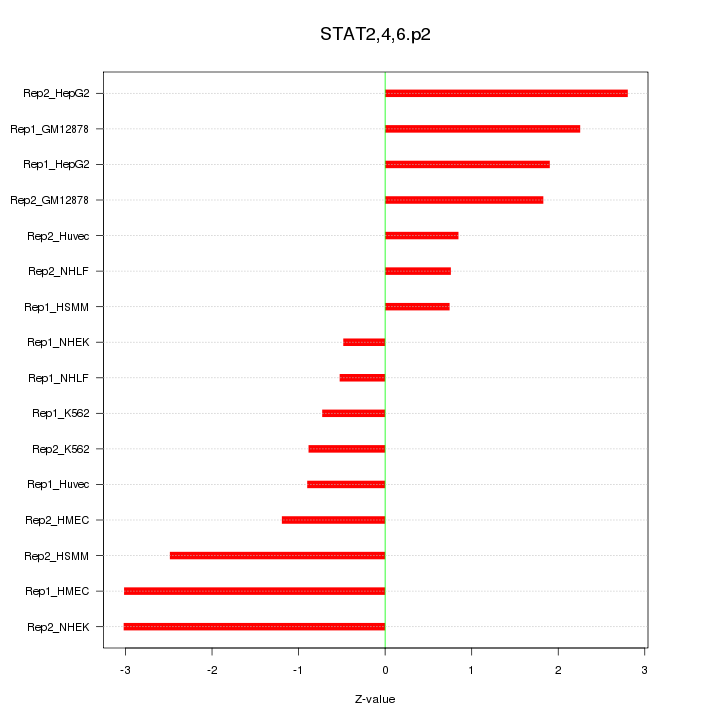
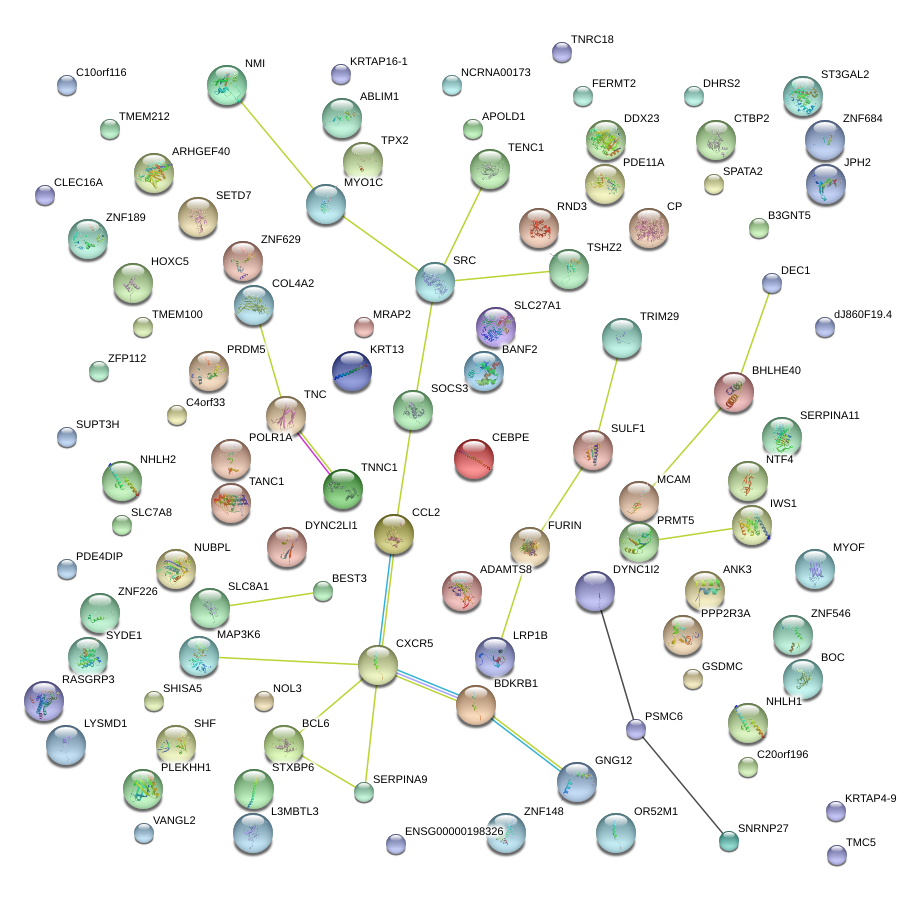
|
chr3_-_188945921
|
3.844
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr17_-_73867748
|
3.540
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr3_-_188946168
|
2.317
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_-_126706442
|
2.249
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr20_+_2743613
|
1.857
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr2_+_69974581
|
1.738
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr2_-_86186441
|
1.711
|
|
POLR1A
|
polymerase (RNA) I polypeptide A, 194kDa
|
|
chr1_-_68071635
|
1.675
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr20_-_48741481
|
1.545
|
|
|
|
|
chr2_-_178645718
|
1.423
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr14_-_22658078
|
1.337
|
NM_001805
|
CEBPE
|
CCAAT/enhancer binding protein (C/EBP), epsilon
|
|
chr9_-_116920232
|
1.318
|
NM_002160
|
TNC
|
tenascin C
|
|
chr17_+_55269974
|
1.315
|
|
|
|
|
chr1_+_158636987
|
1.223
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr20_+_29790564
|
1.221
|
NM_012112
|
TPX2
|
TPX2, microtubule-associated, homolog (Xenopus laevis)
|
|
chr2_+_33554824
|
1.206
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr20_+_17622319
|
1.183
|
NM_001159495
|
BANF2
|
barrier to autointegration factor 2
|
|
chr12_+_12829807
|
1.182
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr1_-_27565923
|
1.158
|
NM_004672
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr10_-_21847126
|
1.141
|
|
|
|
|
chr19_+_45194782
|
1.118
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chr1_+_68070573
|
1.107
|
|
|
|
|
chr13_+_109932934
|
1.106
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr12_+_115455579
|
1.097
|
|
NCRNA00173
|
non-protein coding RNA 173
|
|
chr14_-_93988838
|
1.095
|
NM_001080451
|
SERPINA11
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11
|
|
chr3_+_114414064
|
1.071
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr16_-_69030421
|
1.071
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr12_-_47532171
|
1.070
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr4_+_130236731
|
1.070
|
NM_001099783
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr7_-_27158726
|
1.062
|
|
|
|
|
chr2_+_172252473
|
1.056
|
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr20_+_35406497
|
1.040
|
NM_005417
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr15_-_43280664
|
1.020
|
NM_138356
|
SHF
|
Src homology 2 domain containing F
|
|
chr17_+_36515166
|
1.009
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr11_-_119504726
|
1.008
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr14_+_95792299
|
0.995
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr6_+_130381408
|
0.989
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr10_-_95231933
|
0.984
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr19_-_49552638
|
0.944
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr14_+_67069668
|
0.941
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr2_-_151052391
|
0.938
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr10_-_116382089
|
0.931
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr14_+_23175383
|
0.922
|
NM_005794
NM_182908
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2
|
|
chr16_+_19329557
|
0.921
|
NM_001105248
NM_001105249
|
TMC5
|
transmembrane channel-like 5
|
|
chr3_-_48516486
|
0.919
|
NM_016479
|
SHISA5
|
shisa homolog 5 (Xenopus laevis)
|
|
chr2_+_43854681
|
0.905
|
NM_001193464
NM_015522
NM_016008
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr17_-_51155073
|
0.893
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr19_-_41711009
|
0.891
|
NM_001012756
NM_001166036
NM_001166037
NM_001166038
|
ZNF260
|
zinc finger protein 260
|
|
chr14_-_22468393
|
0.885
|
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr8_+_70567581
|
0.866
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr16_-_30705707
|
0.864
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr14_+_31100330
|
0.864
|
NM_025152
|
NUBPL
|
nucleotide binding protein-like
|
|
chr11_+_4522993
|
0.861
|
NM_001004137
|
OR52M1
|
olfactory receptor, family 52, subfamily M, member 1
|
|
chr14_+_52243645
|
0.860
|
NM_002806
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6
|
|
chr19_-_54258935
|
0.853
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr7_-_5429702
|
0.849
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr20_-_47963682
|
0.844
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr14_-_22722658
|
0.841
|
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr14_+_20608179
|
0.818
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr2_-_40532651
|
0.815
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr8_-_130868315
|
0.806
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr19_+_49361054
|
0.788
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr4_-_140696740
|
0.788
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr6_+_84799969
|
0.786
|
NM_138409
|
MRAP2
|
melanocortin 2 receptor accessory protein 2
|
|
chr9_+_103200939
|
0.786
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr3_+_173043832
|
0.774
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr2_+_159533329
|
0.772
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr14_-_52487022
|
0.771
|
|
FERMT2
|
fermitin family member 2
|
|
chr20_-_42249146
|
0.770
|
|
JPH2
|
junctophilin 2
|
|
chr19_+_15079141
|
0.765
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr20_+_5679032
|
0.764
|
NM_152504
|
C20orf196
|
chromosome 20 open reading frame 196
|
|
chr3_-_150422474
|
0.763
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr14_-_24588916
|
0.760
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr9_+_116943917
|
0.758
|
NM_017418
|
DEC1
|
deleted in esophageal cancer 1
|
|
chr17_-_36915329
|
0.747
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr12_-_68369322
|
0.746
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr17_-_36719030
|
0.745
|
NM_001146182
|
KRTAP16-1
|
keratin associated protein 16-1
|
|
chr2_-_142605739
|
0.743
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr19_+_17440577
|
0.740
|
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr14_-_22722680
|
0.739
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr11_-_118693021
|
0.739
|
NM_006500
|
MCAM
|
melanoma cell adhesion molecule
|
|
chr11_+_118259684
|
0.738
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr6_-_45453552
|
0.728
|
NM_003599
NM_181356
|
SUPT3H
|
suppressor of Ty 3 homolog (S. cerevisiae)
|
|
chr3_+_137224266
|
0.725
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr2_-_128000315
|
0.725
|
NM_017969
|
IWS1
|
IWS1 homolog (S. cerevisiae)
|
|
chr8_+_70567634
|
0.722
|
|
SULF1
|
sulfatase 1
|
|
chr12_+_51730101
|
0.719
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr10_+_88718167
|
0.707
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr15_+_89212888
|
0.706
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr17_+_29606408
|
0.702
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr4_-_122063118
|
0.702
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr16_+_10945815
|
0.700
|
NM_015226
|
CLEC16A
|
C-type lectin domain family 16, member A
|
|
chr3_+_184453547
|
0.698
|
NM_032047
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr1_+_40769819
|
0.694
|
NM_152373
|
ZNF684
|
zinc finger protein 684
|
|
chr10_-_62163083
|
0.686
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_-_22693333
|
0.684
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr3_-_126576693
|
0.677
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr16_+_65761905
|
0.677
|
NM_001185058
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr1_-_116184846
|
0.675
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr1_-_143706300
|
0.673
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_-_151854619
|
0.672
|
NM_004688
|
NMI
|
N-myc (and STAT) interactor
|
|
chr1_-_149404954
|
0.671
|
NM_212551
NM_001136543
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1
|
|
chr20_+_51023159
|
0.667
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr12_+_52713098
|
0.666
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr6_+_26312758
|
0.664
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr18_+_3437607
|
0.664
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr14_-_22468445
|
0.663
|
NM_001039619
NM_006109
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr6_-_29081015
|
0.657
|
NM_001010877
|
ZNF311
|
zinc finger protein 311
|
|
chr14_-_22516271
|
0.656
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr1_+_40769877
|
0.654
|
|
ZNF684
|
zinc finger protein 684
|
|
chr20_+_17628592
|
0.654
|
NM_001014977
NM_178477
|
BANF2
|
barrier to autointegration factor 2
|
|
chr1_+_36462625
|
0.654
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr7_-_87774130
|
0.651
|
NM_024636
|
STEAP4
|
STEAP family member 4
|
|
chr6_-_31797434
|
0.650
|
NM_025261
|
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr16_-_18709125
|
0.647
|
NM_001019
NM_001030009
|
RPS15A
|
ribosomal protein S15a
|
|
chr11_-_85115105
|
0.646
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr20_+_57948803
|
0.640
|
NM_001190827
NM_022106
|
C20orf177
|
chromosome 20 open reading frame 177
|
|
chr2_-_142605010
|
0.637
|
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr11_+_36572615
|
0.636
|
NM_138787
|
C11orf74
|
chromosome 11 open reading frame 74
|
|
chr5_-_180563945
|
0.634
|
NM_203294
NM_203295
NM_203296
|
TRIM7
|
tripartite motif containing 7
|
|
chr14_+_23855291
|
0.633
|
|
LTB4R
|
leukotriene B4 receptor
|
|
chr20_+_54637714
|
0.632
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr17_-_38979178
|
0.629
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr9_-_129729518
|
0.628
|
NM_173492
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1
|
|
chr17_+_11864841
|
0.627
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr12_-_41164052
|
0.622
|
NM_001144882
NM_001144883
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr14_+_19881552
|
0.620
|
NM_001042618
NM_005484
|
PARP2
|
poly (ADP-ribose) polymerase 2
|
|
chr7_+_138469029
|
0.617
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr2_-_61551350
|
0.611
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr21_-_34749908
|
0.607
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr5_+_40877053
|
0.606
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr11_-_86061301
|
0.599
|
NM_001161586
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr14_+_22305570
|
0.599
|
NM_005015
|
OXA1L
|
oxidase (cytochrome c) assembly 1-like
|
|
chr14_+_31868229
|
0.597
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_-_148494732
|
0.594
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr2_-_55500450
|
0.593
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr12_-_41164010
|
0.592
|
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr16_-_15644465
|
0.591
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr12_+_13088412
|
0.590
|
|
KIAA1467
|
KIAA1467
|
|
chr12_+_67366989
|
0.589
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr1_+_32485404
|
0.587
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr16_-_73842884
|
0.584
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr19_+_49309350
|
0.581
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr8_-_10550026
|
0.580
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr1_-_143706265
|
0.572
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr9_-_116305273
|
0.571
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr3_+_189353502
|
0.571
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr20_+_5935897
|
0.567
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr15_+_56217686
|
0.566
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr1_-_143706196
|
0.561
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr16_+_15644624
|
0.558
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr18_-_26876622
|
0.557
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr1_-_225572419
|
0.553
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr14_+_23700261
|
0.550
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr2_+_135526304
|
0.547
|
NM_001172435
NM_012233
|
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic)
|
|
chr17_+_64922420
|
0.547
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr16_+_88515917
|
0.546
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr3_+_50204026
|
0.541
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr20_-_14266200
|
0.541
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr16_+_2461500
|
0.541
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr12_-_642786
|
0.540
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr2_+_108360798
|
0.538
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr19_-_49131250
|
0.537
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr4_-_152901542
|
0.532
|
NM_004564
|
PET112L
|
PET112-like (yeast)
|
|
chr10_+_75343352
|
0.530
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr2_+_27740254
|
0.530
|
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr1_-_151788341
|
0.529
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr15_-_81412354
|
0.527
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr14_-_29466562
|
0.524
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr2_-_56266408
|
0.524
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr2_+_32244406
|
0.521
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr12_+_8866416
|
0.520
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr2_-_227952249
|
0.519
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr8_-_127639892
|
0.518
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr3_-_131948202
|
0.515
|
NM_014602
|
PIK3R4
|
phosphoinositide-3-kinase, regulatory subunit 4
|
|
chr1_-_166372244
|
0.514
|
NM_153832
|
GPR161
|
G protein-coupled receptor 161
|
|
chr11_-_57173823
|
0.513
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr5_+_148631588
|
0.509
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr6_+_46728590
|
0.507
|
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr6_-_91063181
|
0.507
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr11_+_117680504
|
0.503
|
NM_000733
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex)
|
|
chr6_-_36463320
|
0.502
|
NM_016135
|
ETV7
|
ets variant 7
|
|
chr19_-_47438583
|
0.501
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr6_-_116973465
|
0.501
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr2_+_113016236
|
0.498
|
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa
|
|
chr6_-_42218692
|
0.498
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr17_-_36760581
|
0.497
|
NM_004138
|
KRT33A
|
keratin 33A
|
|
chr7_-_22363036
|
0.496
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr6_+_31648071
|
0.494
|
NM_000595
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr17_-_36528113
|
0.493
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr6_-_46997579
|
0.493
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr5_+_148631636
|
0.489
|
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr20_-_20641122
|
0.488
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr11_-_6661093
|
0.485
|
NM_022061
|
MRPL17
|
mitochondrial ribosomal protein L17
|
|
chr20_-_33580861
|
0.483
|
NM_001145350
|
C20orf173
|
chromosome 20 open reading frame 173
|
|
chr6_+_167445284
|
0.482
|
NM_004367
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr20_+_51022352
|
0.482
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr13_-_42464301
|
0.481
|
NM_001002264
NM_033255
|
EPSTI1
|
epithelial stromal interaction 1 (breast)
|
|
chr16_+_22216223
|
0.478
|
NM_018119
|
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD)
|
|
chr9_-_88751923
|
0.477
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|