|
chr22_+_20068039
|
3.384
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr22_-_20235372
|
3.371
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr11_+_70926718
|
3.326
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr7_-_130891861
|
3.016
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr2_+_85834111
|
2.581
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr17_+_36493927
|
2.387
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr9_-_96441409
|
2.322
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr15_+_81567327
|
2.161
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr15_-_53822468
|
2.103
|
NM_173814
|
PRTG
|
protogenin
|
|
chr2_-_241484245
|
2.010
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr13_+_30378311
|
1.962
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr9_-_96441654
|
1.810
|
NM_000507
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr8_+_37773991
|
1.769
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr17_-_36594652
|
1.751
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr17_+_4348877
|
1.738
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr19_+_43446937
|
1.689
|
NM_001166103
NM_021102
|
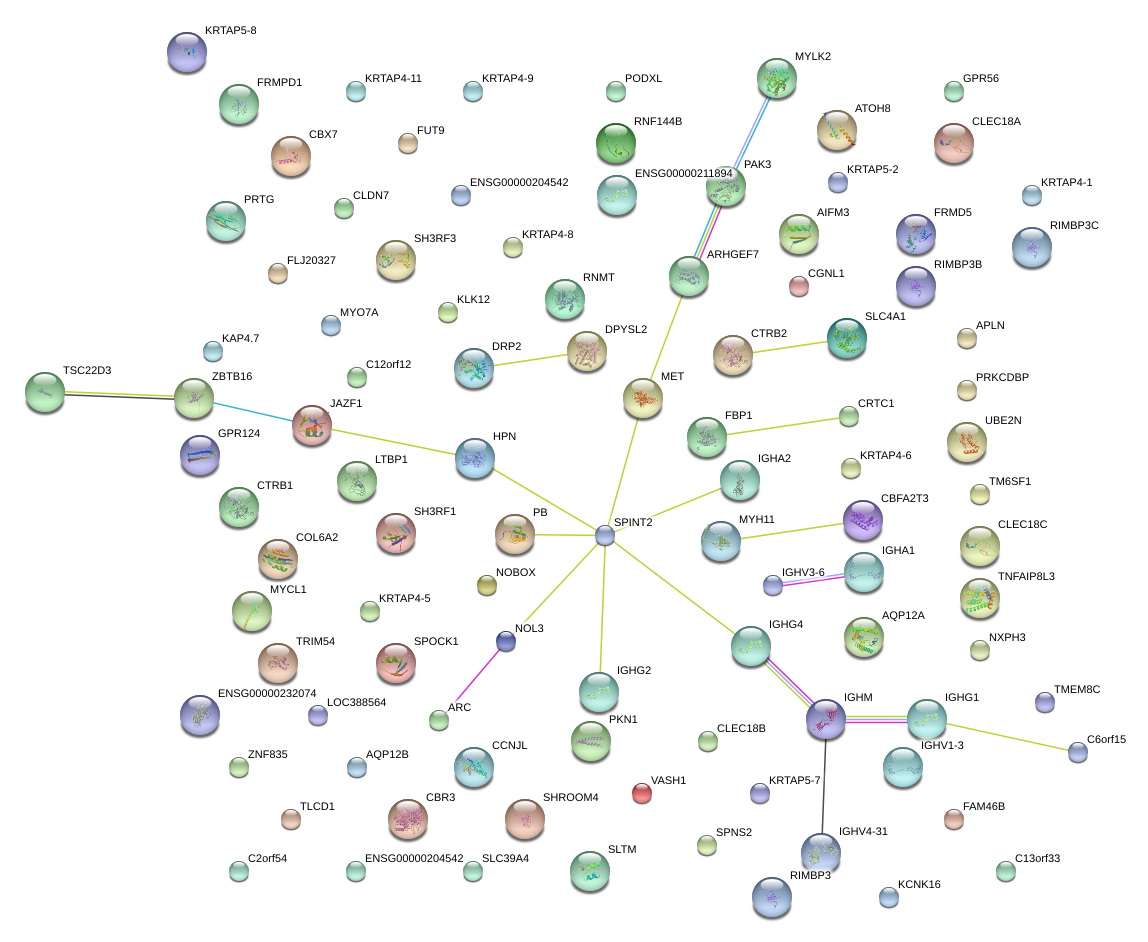
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chrX_-_50573759
|
1.664
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr12_-_89873081
|
1.654
|
NM_152638
|
C12orf12
|
chromosome 12 open reading frame 12
|
|
chr15_-_42274529
|
1.643
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr14_+_76298693
|
1.636
|
|
VASH1
|
vasohibin 1
|
|
chr16_-_73798539
|
1.629
|
NM_001025200
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr21_+_36429079
|
1.602
|
NM_001236
|
CBR3
|
carbonyl reductase 3
|
|
chr19_+_43447078
|
1.598
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_43447262
|
1.597
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_27211913
|
1.551
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr2_-_241270982
|
1.528
|
NM_001102467
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr6_+_18495559
|
1.524
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr1_-_40140126
|
1.495
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr6_+_96570565
|
1.492
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr11_+_113436438
|
1.463
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_143692814
|
1.449
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr2_+_33213111
|
1.430
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr17_-_7106987
|
1.417
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr15_-_42274760
|
1.398
|
|
FRMD5
|
FERM domain containing 5
|
|
chrX_-_13866366
|
1.360
|
|
|
|
|
chr2_+_241279934
|
1.321
|
NM_198998
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr16_-_87570716
|
1.320
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr17_+_45008175
|
1.319
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr9_-_96441567
|
1.319
|
|
FBP1
|
fructose-1,6-bisphosphatase 1
|
|
chr22_+_19660361
|
1.316
|
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr17_+_36515166
|
1.292
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr11_+_76516957
|
1.290
|
NM_000260
NM_001127179
NM_001127180
|
MYO7A
|
myosin VIIA
|
|
chr7_-_143731718
|
1.286
|
NM_001080413
|
NOBOX
|
NOBOX oogenesis homeobox
|
|
chr19_+_18655487
|
1.257
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr11_-_6298284
|
1.254
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chrX_+_110074124
|
1.254
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr15_+_55455994
|
1.249
|
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr5_-_159672060
|
1.244
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chrX_-_128616594
|
1.234
|
NM_017413
|
APLN
|
apelin
|
|
chr2_+_27358800
|
1.206
|
NM_032546
NM_187841
|
TRIM54
|
tripartite motif containing 54
|
|
chrX_-_106905545
|
1.199
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr17_-_36507900
|
1.198
|
NM_031960
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chrX_+_100361588
|
1.188
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr7_-_28186867
|
1.185
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr16_+_56230707
|
1.181
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr2_+_109112428
|
1.166
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr21_+_46355797
|
1.152
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr19_-_61874934
|
1.151
|
|
ZNF835
|
zinc finger protein 835
|
|
chr22_-_37858871
|
1.151
|
|
CBX7
|
chromobox homolog 7
|
|
chr6_-_39398293
|
1.138
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr9_+_37640996
|
1.134
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr17_-_39700960
|
1.127
|
NM_000342
|
SLC4A1
|
solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group)
|
|
chr19_-_60587414
|
1.119
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr17_-_24077246
|
1.115
|
NM_138463
|
TLCD1
|
TLC domain containing 1
|
|
chr8_-_145609737
|
1.110
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr11_+_70915960
|
1.103
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr16_+_68765428
|
1.102
|
NM_173619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr19_+_40224480
|
1.098
|
|
HPN
|
hepsin
|
|
chr19_-_56229808
|
1.095
|
NM_019598
NM_145894
NM_145895
|
KLK12
|
kallikrein-related peptidase 12
|
|
chr5_-_136862130
|
1.093
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr15_-_49184764
|
1.088
|
NM_207381
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3
|
|
chr1_+_181259317
|
1.088
|
|
|
|
|
chr20_+_29870836
|
1.085
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr7_+_116099648
|
1.069
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr1_-_27212035
|
1.064
|
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr19_+_14405216
|
1.059
|
|
PKN1
|
protein kinase N1
|
|
chr16_-_15742883
|
1.057
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr14_-_105948734
|
1.054
|
|
LOC100126583
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr17_-_36559579
|
1.042
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr9_-_135379888
|
1.041
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr6_-_31188310
|
1.039
|
NM_014070
|
C6orf15
|
chromosome 6 open reading frame 15
|
|
chrX_-_106905657
|
1.039
|
NM_198057
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr19_-_51829778
|
1.039
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr16_+_68542310
|
1.038
|
NM_182619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chrX_+_48221323
|
1.037
|
NM_177434
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr17_+_36599664
|
1.036
|
NM_001190460
|
KRTAP9-1
|
keratin associated protein 9-1
|
|
chrX_-_13866451
|
1.032
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_199975271
|
1.030
|
|
NAV1
|
neuron navigator 1
|
|
chr11_+_61204392
|
1.029
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr11_+_1816808
|
1.029
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr5_+_169465478
|
1.025
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr17_+_37694007
|
1.024
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr1_+_100776280
|
1.020
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr8_-_139578246
|
1.019
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr1_-_6585268
|
1.004
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr1_+_24614831
|
1.002
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr6_-_31977747
|
1.002
|
NM_181842
|
ZBTB12
|
zinc finger and BTB domain containing 12
|
|
chr15_-_88143721
|
0.995
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr9_+_135389698
|
0.986
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr12_+_108690199
|
0.985
|
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr4_+_151218862
|
0.969
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr12_+_47498778
|
0.968
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr7_-_120285245
|
0.967
|
|
TSPAN12
|
tetraspanin 12
|
|
chr15_-_58671929
|
0.962
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr17_+_26060617
|
0.962
|
|
|
|
|
chr17_-_7106497
|
0.948
|
|
CLDN7
|
claudin 7
|
|
chr11_-_16992524
|
0.944
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr15_-_72981595
|
0.943
|
|
C15orf17
|
chromosome 15 open reading frame 17
|
|
chr1_-_39871226
|
0.943
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr7_-_135083906
|
0.940
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr14_+_76297885
|
0.939
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr3_+_32255215
|
0.929
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr2_+_233099113
|
0.918
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr1_+_201364050
|
0.915
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr3_+_32255158
|
0.914
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr2_-_101134146
|
0.912
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr16_+_87232501
|
0.912
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr22_+_29122932
|
0.911
|
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr11_-_684842
|
0.906
|
|
DEAF1
|
deformed epidermal autoregulatory factor 1 (Drosophila)
|
|
chr11_+_817584
|
0.904
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr5_+_66160359
|
0.904
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr6_+_36754467
|
0.899
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr3_+_141879555
|
0.889
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr19_+_40224457
|
0.887
|
|
HPN
|
hepsin
|
|
chr6_+_7672009
|
0.886
|
NM_001718
|
BMP6
|
bone morphogenetic protein 6
|
|
chr16_+_971808
|
0.880
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr10_-_21847126
|
0.879
|
|
|
|
|
chr5_+_159276317
|
0.866
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr2_+_128119908
|
0.866
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr10_-_61819493
|
0.865
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr9_+_123544884
|
0.865
|
NM_138709
|
DAB2IP
|
DAB2 interacting protein
|
|
chr5_-_136862317
|
0.862
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr11_-_65070625
|
0.859
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr12_+_129388543
|
0.859
|
|
PIWIL1
|
piwi-like 1 (Drosophila)
|
|
chr12_+_103675075
|
0.857
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr22_-_30071761
|
0.851
|
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr2_-_80384454
|
0.850
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr2_+_176761552
|
0.848
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr17_+_36665161
|
0.846
|
NM_030975
|
KRTAP9-9
|
keratin associated protein 9-9
|
|
chr9_-_129869211
|
0.845
|
NM_197956
|
NAIF1
|
nuclear apoptosis inducing factor 1
|
|
chr19_+_18655483
|
0.841
|
|
CRTC1
|
CREB regulated transcription coactivator 1
|
|
chr16_-_73012740
|
0.840
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chrX_+_53095230
|
0.833
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr8_-_131483395
|
0.832
|
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr5_+_1254709
|
0.827
|
NM_001003841
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19
|
|
chr5_+_71438796
|
0.823
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_-_1026654
|
0.822
|
NM_005961
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|
|
chr1_+_199975674
|
0.820
|
|
NAV1
|
neuron navigator 1
|
|
chr17_+_36636404
|
0.816
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr20_+_42007949
|
0.813
|
NM_001098798
|
TOX2
|
TOX high mobility group box family member 2
|
|
chr17_-_70220605
|
0.809
|
NM_139018
|
CD300LF
|
CD300 molecule-like family member f
|
|
chr5_+_143171918
|
0.808
|
NM_021182
|
HMHB1
|
histocompatibility (minor) HB-1
|
|
chr15_-_23659329
|
0.803
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr19_-_59495949
|
0.799
|
NM_001172654
NM_006865
|
LILRB2
LILRA3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr1_+_18680010
|
0.797
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr16_+_972064
|
0.795
|
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr12_+_6819635
|
0.793
|
NM_002075
|
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3
|
|
chr22_-_36212160
|
0.791
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr6_+_138524717
|
0.790
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chrX_+_69559605
|
0.787
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr13_+_112789669
|
0.786
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_+_18081968
|
0.785
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr2_+_56265260
|
0.782
|
|
|
|
|
chr6_+_167658563
|
0.779
|
NM_031949
|
TTLL2
|
tubulin tyrosine ligase-like family, member 2
|
|
chr5_+_148501178
|
0.779
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr19_+_51498695
|
0.765
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr14_-_105851580
|
0.764
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_130980359
|
0.762
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr2_-_183439515
|
0.762
|
NM_001463
|
FRZB
|
frizzled-related protein
|
|
chrX_+_70438352
|
0.757
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr9_-_139060285
|
0.756
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr16_+_6473759
|
0.756
|
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_+_45488330
|
0.754
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_-_23729871
|
0.748
|
|
E2F2
|
E2F transcription factor 2
|
|
chr4_-_191143017
|
0.747
|
NM_020040
|
TUBB4Q
|
tubulin, beta polypeptide 4, member Q
|
|
chr3_+_32123140
|
0.744
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr2_-_216944911
|
0.742
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr15_+_66658626
|
0.740
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr8_+_31010319
|
0.740
|
NM_000553
|
WRN
|
Werner syndrome, RecQ helicase-like
|
|
chr1_+_46418786
|
0.738
|
|
TSPAN1
|
tetraspanin 1
|
|
chr1_+_242281175
|
0.735
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr9_-_115101140
|
0.732
|
NM_145051
|
RNF183
|
ring finger protein 183
|
|
chr11_+_113435640
|
0.731
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr12_-_68379331
|
0.729
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr1_-_6585485
|
0.728
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr10_-_116382089
|
0.727
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_1104939
|
0.727
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr8_-_49996353
|
0.722
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr8_+_145138874
|
0.721
|
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
|
|
chr17_+_45488356
|
0.721
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_+_199975563
|
0.721
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr16_-_71549149
|
0.719
|
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr11_-_63689780
|
0.716
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr19_-_59438411
|
0.715
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr14_-_105549177
|
0.714
|
|
IGHV4-31
IGHD
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr19_-_2259155
|
0.713
|
NM_001101391
|
LINGO3
|
leucine rich repeat and Ig domain containing 3
|
|
chr17_+_40192072
|
0.712
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr2_+_10101081
|
0.711
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr12_+_48630790
|
0.708
|
NM_000486
|
AQP2
|
aquaporin 2 (collecting duct)
|
|
chr9_-_139060377
|
0.707
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|