|
chr5_-_22248630
|
1.204
|
|
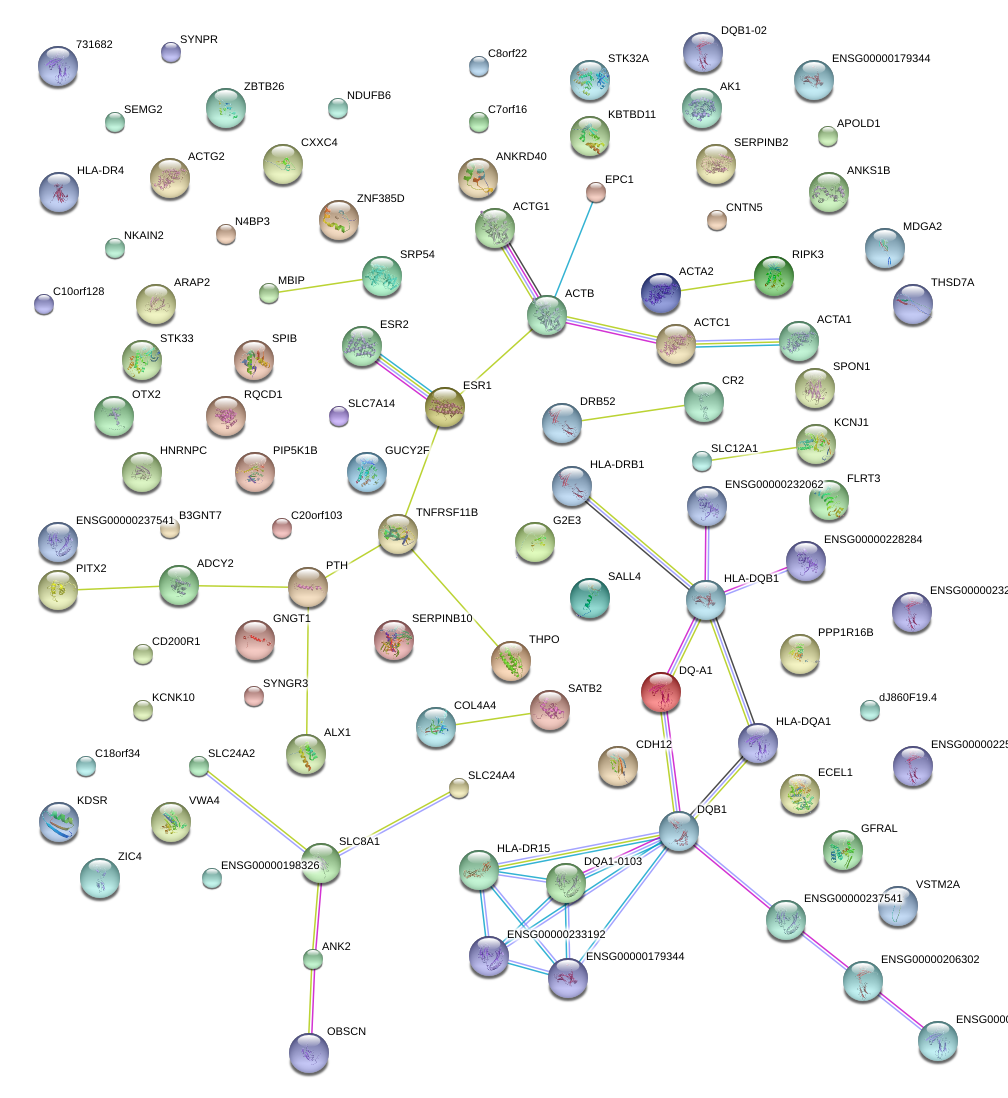
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr2_-_200038055
|
1.134
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr12_+_84198015
|
1.117
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr11_-_8571988
|
1.009
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr3_+_131547048
|
0.948
|
NM_153264
|
COL6A5
|
collagen, type VI, alpha 5
|
|
chr1_+_205694237
|
0.933
|
NM_001006658
NM_001877
|
CR2
|
complement component (3d/Epstein Barr virus) receptor 2
|
|
chr7_+_54577512
|
0.889
|
NM_182546
|
VSTM2A
|
V-set and transmembrane domain containing 2A
|
|
chr4_+_114190233
|
0.887
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr10_-_50066386
|
0.874
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr7_+_31693155
|
0.865
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr11_-_13474101
|
0.846
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr6_+_124166767
|
0.815
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr14_-_20807346
|
0.805
|
NM_001077442
NM_001077443
NM_004500
NM_031314
|
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2)
|
|
chr6_-_32742265
|
0.800
|
NM_002123
|
HLA-DQB1
|
major histocompatibility complex, class II, DQ beta 1
|
|
chr20_+_43283423
|
0.778
|
NM_003008
|
SEMG2
|
semenogelin II
|
|
chr15_+_46285789
|
0.766
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr8_-_120033240
|
0.751
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr2_-_233060774
|
0.750
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr20_+_36867708
|
0.746
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr14_-_47213906
|
0.744
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr3_-_114176401
|
0.738
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr10_-_32707549
|
0.715
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr2_-_104834344
|
0.714
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr2_-_227737009
|
0.702
|
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr3_-_185578621
|
0.694
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr2_-_40593074
|
0.685
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr10_-_90702490
|
0.679
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr3_-_171786468
|
0.675
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr14_-_56342097
|
0.674
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr7_+_93373755
|
0.660
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr16_+_1979959
|
0.648
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr2_+_231968578
|
0.645
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr11_-_128217550
|
0.643
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr2_+_219141761
|
0.633
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr6_+_152168500
|
0.626
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr7_-_11838260
|
0.622
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr3_+_63403792
|
0.622
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr3_-_148607096
|
0.613
|
NM_032153
|
ZIC4
|
Zic family member 4
|
|
chr20_-_14266200
|
0.610
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr11_+_98396789
|
0.607
|
NM_014361
NM_175566
|
CNTN5
|
contactin 5
|
|
chr14_-_35859545
|
0.605
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr9_+_70509925
|
0.605
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr3_-_22389126
|
0.604
|
|
ZNF385D
|
zinc finger protein 385D
|
|
chr9_-_32563057
|
0.601
|
NM_002493
NM_182739
|
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa
|
|
chr14_+_34521854
|
0.593
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr11_-_8572277
|
0.589
|
|
STK33
|
serine/threonine kinase 33
|
|
chr14_+_30098079
|
0.581
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr14_+_34521945
|
0.580
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr4_-_105635499
|
0.575
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr5_+_146594718
|
0.573
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr6_+_55300225
|
0.569
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr4_-_35922288
|
0.563
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr14_-_23878871
|
0.560
|
NM_006871
|
RIPK3
|
receptor-interacting serine-threonine kinase 3
|
|
chr5_+_177473071
|
0.556
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr14_-_87862676
|
0.555
|
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr14_-_87863003
|
0.552
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_+_13940715
|
0.549
|
|
SPON1
|
spondin 1, extracellular matrix protein
|
|
chr20_-_604443
|
0.546
|
|
|
|
|
chr17_-_46140015
|
0.534
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr18_-_59185330
|
0.531
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr12_+_12829807
|
0.531
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr18_-_29273979
|
0.524
|
NM_001105528
|
C18orf34
|
chromosome 18 open reading frame 34
|
|
chr12_-_97812633
|
0.524
|
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr4_-_111777584
|
0.517
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr20_-_49852353
|
0.514
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr19_+_55614006
|
0.513
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr6_+_131190237
|
0.512
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr6_+_32713160
|
0.511
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr8_+_50147455
|
0.506
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr9_-_124733599
|
0.501
|
NM_020924
|
ZBTB26
|
zinc finger and BTB domain containing 26
|
|
chr5_+_7449030
|
0.497
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr20_+_2743613
|
0.492
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr8_+_1940282
|
0.486
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chrX_-_108611940
|
0.485
|
NM_001522
|
GUCY2F
|
guanylate cyclase 2F, retinal
|
|
chr20_+_9442995
|
0.484
|
|
C20orf103
|
chromosome 20 open reading frame 103
|
|
chr18_+_59733724
|
0.481
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr9_-_19778590
|
0.480
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr11_+_122901689
|
0.479
|
NM_020716
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr8_+_55210333
|
0.476
|
NM_014175
|
MRPL15
|
mitochondrial ribosomal protein L15
|
|
chr21_-_38955487
|
0.474
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr10_-_115923874
|
0.473
|
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr11_-_8572321
|
0.473
|
|
STK33
|
serine/threonine kinase 33
|
|
chr15_-_24569201
|
0.472
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr11_-_19180092
|
0.470
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr12_-_21985433
|
0.468
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr17_+_43165608
|
0.468
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr17_-_44010543
|
0.466
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr5_+_5193442
|
0.464
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr3_-_166397142
|
0.462
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr2_-_122210873
|
0.453
|
NM_032390
|
MKI67IP
|
MKI67 (FHA domain) interacting nucleolar phosphoprotein
|
|
chr1_+_57093030
|
0.448
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr12_+_79995927
|
0.446
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr1_-_68688219
|
0.446
|
NM_000329
|
RPE65
|
retinal pigment epithelium-specific protein 65kDa
|
|
chr13_-_46368968
|
0.443
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr3_+_87359088
|
0.441
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr9_-_78710698
|
0.441
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr10_+_88506320
|
0.438
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr3_+_88114415
|
0.436
|
NM_000866
|
HTR1F
|
5-hydroxytryptamine (serotonin) receptor 1F
|
|
chrX_+_151557292
|
0.436
|
NM_018558
|
GABRQ
|
gamma-aminobutyric acid (GABA) receptor, theta
|
|
chr2_-_74424041
|
0.435
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chrX_+_144707022
|
0.430
|
NM_001144003
NM_001144004
NM_001144005
NM_032539
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr16_-_87535106
|
0.430
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_+_73810383
|
0.426
|
NM_001906
|
CTRB1
CTRB2
|
chymotrypsinogen B1
chymotrypsinogen B2
|
|
chr6_-_32665409
|
0.425
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr14_+_34585098
|
0.422
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr14_-_36059084
|
0.419
|
NM_001079668
|
NKX2-1
|
NK2 homeobox 1
|
|
chr17_-_39375327
|
0.418
|
NM_002722
|
PPY
|
pancreatic polypeptide
|
|
chr6_+_54819527
|
0.416
|
NM_001010872
|
FAM83B
|
family with sequence similarity 83, member B
|
|
chr10_-_134449467
|
0.416
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chrX_+_23262905
|
0.415
|
NM_173495
|
PTCHD1
|
patched domain containing 1
|
|
chr4_-_111777651
|
0.412
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr19_-_6671585
|
0.411
|
NM_000064
|
C3
|
complement component 3
|
|
chr20_+_5839973
|
0.411
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr21_-_36870736
|
0.409
|
NM_001146077
|
CLDN14
|
claudin 14
|
|
chr3_+_123279439
|
0.407
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr13_+_87122778
|
0.407
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr2_-_210798232
|
0.407
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr20_-_13713509
|
0.407
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr3_-_99236520
|
0.406
|
NM_001105580
|
GABRR3
|
gamma-aminobutyric acid (GABA) receptor, rho 3
|
|
chr18_+_27332024
|
0.406
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr3_-_53900888
|
0.405
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr20_+_43269051
|
0.403
|
NM_003007
|
SEMG1
|
semenogelin I
|
|
chr22_+_40286957
|
0.403
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr6_-_52736231
|
0.402
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr1_+_195123766
|
0.401
|
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr7_-_32076985
|
0.401
|
NM_001191056
NM_001191057
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr10_+_118295417
|
0.399
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr10_+_96295511
|
0.399
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr15_-_24569309
|
0.394
|
NM_000814
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr17_+_55269974
|
0.394
|
|
|
|
|
chr4_-_145281237
|
0.393
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr17_-_63798851
|
0.390
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr11_-_95715976
|
0.390
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr10_-_12124778
|
0.389
|
NM_015542
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr15_+_56511466
|
0.389
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr18_+_48121067
|
0.388
|
|
DCC
|
deleted in colorectal carcinoma
|
|
chr1_-_79244948
|
0.388
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr3_-_120236118
|
0.385
|
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr11_+_8660913
|
0.384
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr20_-_61462960
|
0.381
|
NM_000744
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4
|
|
chr21_+_33320080
|
0.381
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr11_-_127962662
|
0.380
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_+_50348052
|
0.376
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr11_+_66952462
|
0.374
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr3_+_150330060
|
0.371
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr17_-_57497380
|
0.371
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr2_+_223433919
|
0.369
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr8_-_93144039
|
0.369
|
|
|
|
|
chr6_+_132901119
|
0.368
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr10_-_7748811
|
0.368
|
NM_001001851
NM_030569
|
ITIH5
|
inter-alpha (globulin) inhibitor H5
|
|
chr2_-_100087366
|
0.367
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr3_+_144377822
|
0.365
|
|
|
|
|
chr12_-_18134319
|
0.364
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr5_-_27074407
|
0.364
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr3_+_116824840
|
0.363
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr15_-_24569253
|
0.362
|
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr1_+_159817752
|
0.361
|
NM_201563
|
FCGR2C
|
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr10_+_102212787
|
0.361
|
NM_003393
|
WNT8B
|
wingless-type MMTV integration site family, member 8B
|
|
chr7_-_31347032
|
0.360
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr10_-_104587069
|
0.357
|
NM_000102
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1
|
|
chr5_+_147238466
|
0.357
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr14_-_19871267
|
0.356
|
NM_021178
NM_182849
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr21_-_38954433
|
0.354
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr14_+_61653827
|
0.352
|
|
FLJ43390
|
hypothetical LOC646113
|
|
chr16_+_1323627
|
0.351
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr2_-_206659140
|
0.351
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr8_+_57520915
|
0.351
|
|
|
|
|
chrX_+_117364444
|
0.351
|
|
WDR44
|
WD repeat domain 44
|
|
chr2_+_231629839
|
0.351
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr4_+_166468266
|
0.350
|
NM_001017369
NM_006745
|
SC4MOL
|
sterol-C4-methyl oxidase-like
|
|
chr6_-_32839244
|
0.350
|
NM_001198858
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr10_-_7491204
|
0.349
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr11_-_19038803
|
0.348
|
NM_054030
|
MRGPRX2
|
MAS-related GPR, member X2
|
|
chr18_+_61569136
|
0.348
|
NM_004361
|
CDH7
|
cadherin 7, type 2
|
|
chr11_-_101828563
|
0.346
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr1_-_245237977
|
0.343
|
NM_020394
|
ZNF695
|
zinc finger protein 695
|
|
chr1_+_18306823
|
0.343
|
NM_032880
|
IGSF21
|
immunoglobin superfamily, member 21
|
|
chr2_+_190753833
|
0.342
|
NM_001042519
NM_032321
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr1_-_120737416
|
0.342
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr16_-_66827449
|
0.342
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr2_-_218551786
|
0.342
|
|
TNS1
|
tensin 1
|
|
chr7_+_50318801
|
0.342
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr20_+_59260847
|
0.340
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr8_+_120497732
|
0.340
|
NM_002514
|
NOV
|
nephroblastoma overexpressed gene
|
|
chr2_+_201099093
|
0.339
|
NM_001160033
NM_001160046
NM_152524
|
SGOL2
|
shugoshin-like 2 (S. pombe)
|
|
chr1_+_195179533
|
0.338
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr18_+_50139168
|
0.338
|
NM_173529
|
C18orf54
|
chromosome 18 open reading frame 54
|
|
chr14_-_22825072
|
0.334
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr3_-_114898003
|
0.333
|
NM_001009899
|
KIAA2018
|
KIAA2018
|
|
chr16_-_3082840
|
0.333
|
NM_032805
|
ZSCAN10
|
zinc finger and SCAN domain containing 10
|
|
chr12_-_16650687
|
0.332
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_190153711
|
0.331
|
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr11_-_1026654
|
0.331
|
NM_005961
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|
|
chr17_+_51585834
|
0.329
|
NM_153228
|
ANKFN1
|
ankyrin-repeat and fibronectin type III domain containing 1
|
|
chr8_+_18111894
|
0.328
|
NM_000662
NM_001160170
NM_001160171
NM_001160172
NM_001160173
NM_001160175
NM_001160176
|
NAT1
|
N-acetyltransferase 1 (arylamine N-acetyltransferase)
|
|
chr1_+_159899528
|
0.328
|
NM_001002273
NM_001002274
NM_001002275
NM_001190828
NM_004001
|
FCGR2B
FCGR2C
|
Fc fragment of IgG, low affinity IIb, receptor (CD32)
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene)
|
|
chr11_-_29995063
|
0.328
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr1_-_188713324
|
0.328
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr12_-_16650697
|
0.327
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_109730858
|
0.327
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|