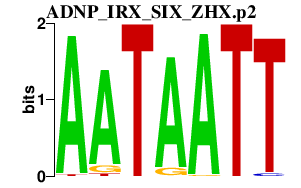
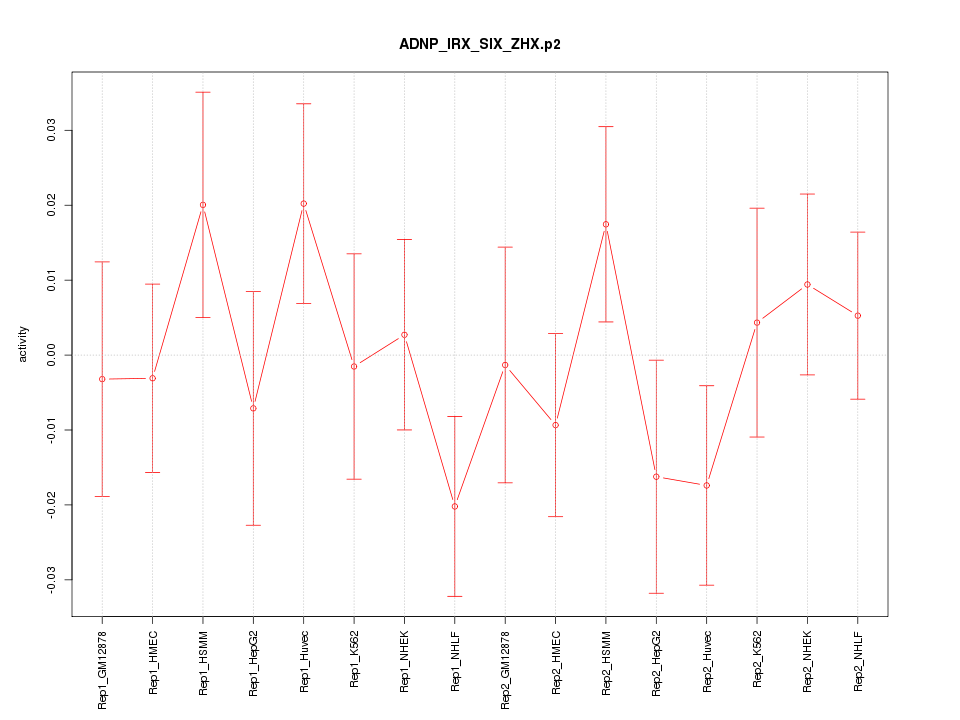
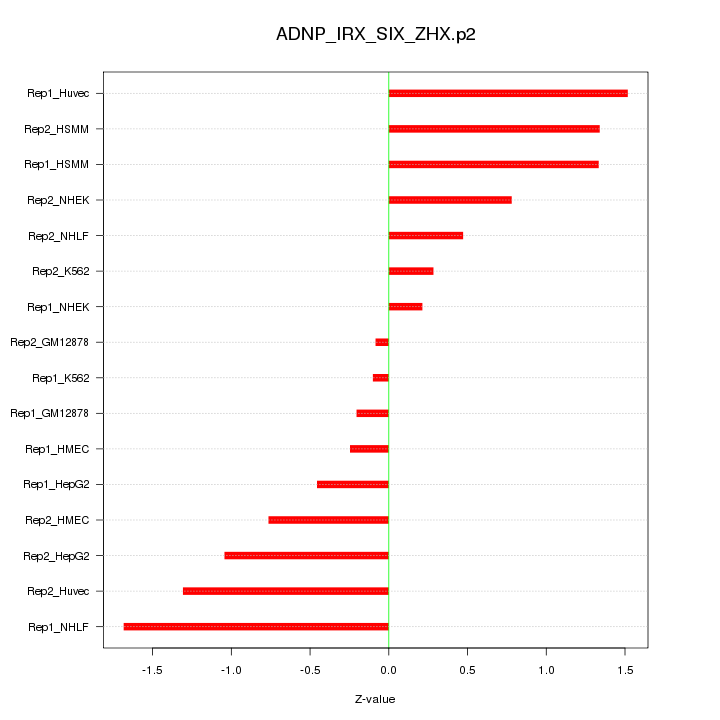
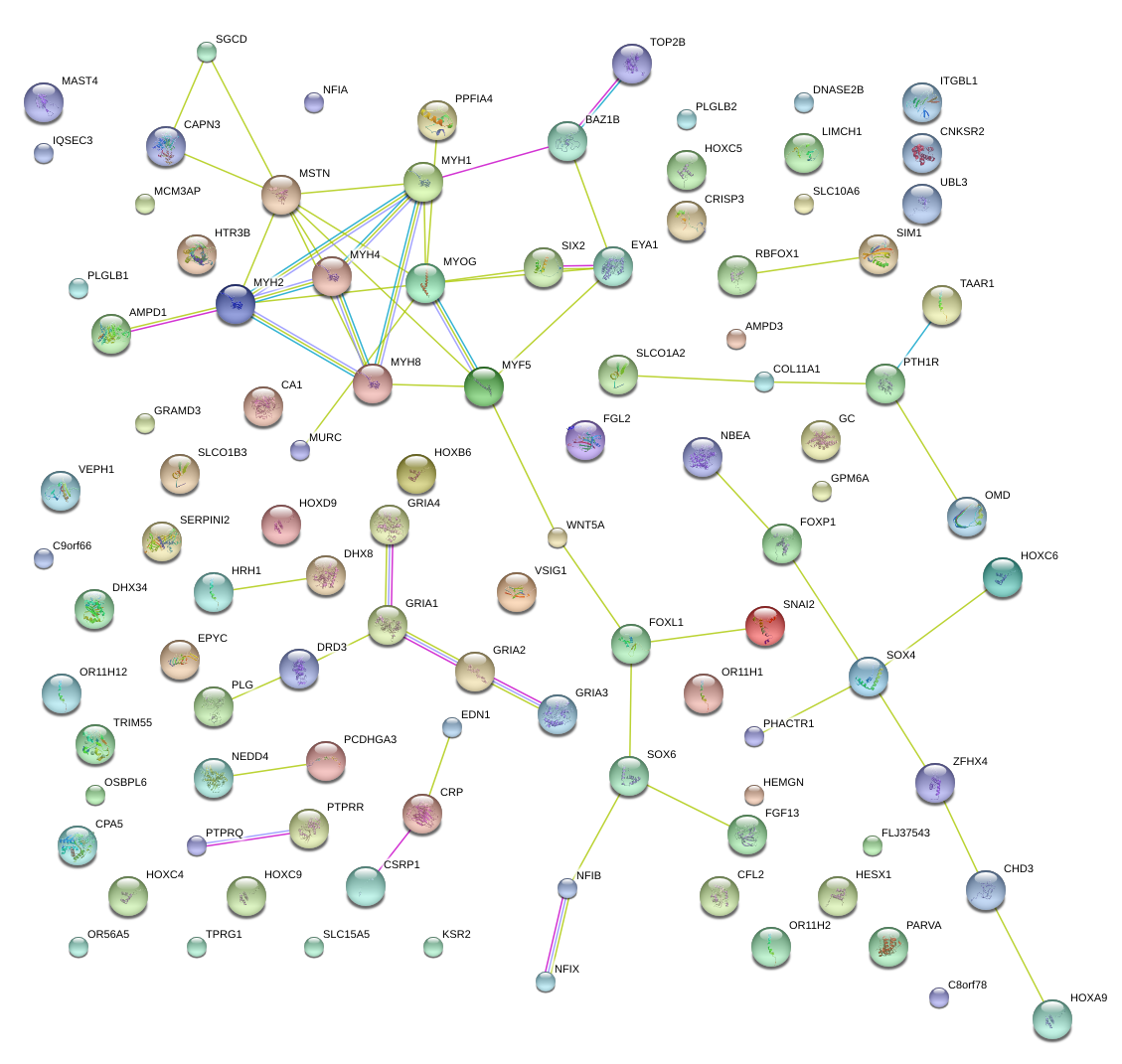
|
chr12_+_52697160
|
1.296
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr7_-_27171673
|
1.291
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr8_+_77756061
|
1.166
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr3_-_168674502
|
1.153
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr7_-_27171648
|
1.096
|
|
HOXA9
|
homeobox A9
|
|
chr8_+_77756077
|
1.093
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr16_+_7322751
|
1.057
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr3_-_55489973
|
1.055
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_-_27171600
|
0.990
|
|
HOXA9
|
homeobox A9
|
|
chr5_+_125786975
|
0.984
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr12_+_52696908
|
0.976
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr9_+_102380156
|
0.817
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr7_-_76667045
|
0.750
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr15_+_40427592
|
0.713
|
NM_212465
|
CAPN3
|
calpain 3, (p94)
|
|
chr8_-_72431274
|
0.704
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr13_+_100902942
|
0.693
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr2_-_45090025
|
0.665
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr9_-_205892
|
0.665
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr17_-_10265991
|
0.659
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr4_-_72868598
|
0.648
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr9_-_14076901
|
0.606
|
|
NFIB
|
nuclear factor I/B
|
|
chr12_+_52680143
|
0.599
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr15_-_53996597
|
0.584
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr5_+_155686333
|
0.576
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr6_+_21701942
|
0.570
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr8_-_72436519
|
0.556
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_+_41309668
|
0.533
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr16_+_85169615
|
0.520
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr4_-_87989291
|
0.517
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr6_-_101018271
|
0.498
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr11_-_5946299
|
0.497
|
NM_001146033
|
OR56A5
|
olfactory receptor, family 56, subfamily A, member 5
|
|
chr8_-_49996353
|
0.494
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr12_-_89922888
|
0.463
|
NM_004950
|
EPYC
|
epiphycan
|
|
chrX_+_107174855
|
0.457
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr8_-_125252884
|
0.442
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr2_+_200031333
|
0.436
|
|
|
|
|
chr13_-_29322795
|
0.435
|
NM_007106
|
UBL3
|
ubiquitin-like 3
|
|
chr1_+_201286933
|
0.435
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr14_-_19252330
|
0.427
|
NM_001197287
|
OR11H2
|
olfactory receptor, family 11, subfamily H, member 2
|
|
chr12_-_21379098
|
0.427
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr3_+_46894239
|
0.426
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr11_+_10433241
|
0.424
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr11_+_113280727
|
0.421
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr3_-_115380539
|
0.419
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr3_-_57209319
|
0.413
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr12_-_116890410
|
0.403
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr7_+_129771865
|
0.397
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr17_-_44037332
|
0.388
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr6_-_133008834
|
0.387
|
NM_138327
|
TAAR1
|
trace amine associated receptor 1
|
|
chr6_+_12825818
|
0.385
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr5_+_125786717
|
0.379
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chrX_-_137649180
|
0.379
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_157950898
|
0.378
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr4_+_41309482
|
0.375
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr21_+_24722913
|
0.373
|
|
FLJ42200
|
FLJ42200 protein
|
|
chr17_-_10393664
|
0.371
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr12_-_16321885
|
0.371
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chrX_-_153698930
|
0.367
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr12_+_79634820
|
0.363
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr6_-_49820030
|
0.360
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr4_+_41309690
|
0.357
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_-_25654659
|
0.356
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr3_+_11242666
|
0.356
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr2_-_190635566
|
0.351
|
NM_005259
|
MSTN
|
myostatin
|
|
chr2_-_87102453
|
0.351
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr12_+_79362256
|
0.347
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr11_+_12355600
|
0.347
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr4_-_176970407
|
0.343
|
|
GPM6A
|
glycoprotein M6A
|
|
chr5_+_66160359
|
0.343
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr12_+_20854904
|
0.343
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr12_-_69434330
|
0.338
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr9_-_99740243
|
0.337
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr8_+_67201684
|
0.335
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr5_+_140772691
|
0.330
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr2_+_178893216
|
0.330
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr3_-_158704056
|
0.328
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr3_+_190147633
|
0.328
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr14_-_34252695
|
0.326
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr22_+_44828338
|
0.326
|
|
|
|
|
chr13_+_34948977
|
0.326
|
|
NBEA
|
neurobeachin
|
|
chr11_-_16454493
|
0.320
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_-_201321999
|
0.319
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr5_+_60969392
|
0.318
|
NM_173667
|
FLJ37543
|
hypothetical protein FLJ37543
|
|
chr17_+_7728819
|
0.314
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr3_-_71625318
|
0.312
|
|
FOXP1
|
forkhead box P1
|
|
chr12_-_89922875
|
0.310
|
|
EPYC
|
epiphycan
|
|
chr8_-_86441115
|
0.301
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr6_+_12398514
|
0.300
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr9_-_94226380
|
0.297
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr12_+_56802
|
0.292
|
NM_015232
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr4_-_176970372
|
0.292
|
|
GPM6A
|
glycoprotein M6A
|
|
chr1_-_115039614
|
0.292
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr5_+_152850276
|
0.292
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr1_-_103269356
|
0.291
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr1_+_84646621
|
0.291
|
NM_058248
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr7_-_72574528
|
0.291
|
NM_032408
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr21_-_46529663
|
0.289
|
NM_003906
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein
|
|
chr3_+_123256902
|
0.288
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr5_+_38481392
|
0.288
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr7_-_72574474
|
0.287
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr2_-_1905508
|
0.287
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr11_+_124539768
|
0.287
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr15_-_48434677
|
0.284
|
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1
|
|
chr15_-_70385709
|
0.284
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr12_-_14024187
|
0.283
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr11_-_83071084
|
0.282
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_-_85975263
|
0.281
|
NM_001017924
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2
|
|
chr10_+_44726635
|
0.278
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr6_+_157141565
|
0.278
|
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr3_+_190831909
|
0.275
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr6_-_52060327
|
0.274
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr6_+_158653679
|
0.270
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr21_-_35343456
|
0.268
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr21_-_35343428
|
0.267
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_73911477
|
0.266
|
|
STAMBP
|
STAM binding protein
|
|
chr12_-_6356783
|
0.265
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr4_+_169250281
|
0.264
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr21_-_35343448
|
0.263
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_53972464
|
0.261
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chrX_+_85404626
|
0.260
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr15_-_48434567
|
0.259
|
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1
|
|
chr11_+_133444029
|
0.256
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chrX_-_18600108
|
0.255
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr21_+_29439768
|
0.255
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr1_+_86707113
|
0.251
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr10_+_75343352
|
0.250
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr12_-_10497195
|
0.249
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr2_-_207291364
|
0.248
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr2_+_171316462
|
0.248
|
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa
|
|
chr2_+_102401580
|
0.247
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chrX_+_144716619
|
0.246
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr12_-_4358968
|
0.246
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr12_+_40117751
|
0.243
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr15_-_48434641
|
0.243
|
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1
|
|
chr12_-_69434650
|
0.242
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr7_-_112514990
|
0.242
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr12_-_101398503
|
0.241
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr7_-_72574383
|
0.240
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr10_-_17211795
|
0.240
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr2_-_200030944
|
0.237
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr17_-_10313600
|
0.236
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chrX_+_146941222
|
0.235
|
|
FTH1P8
|
ferritin, heavy polypeptide 1 pseudogene 8
|
|
chr8_-_37847095
|
0.233
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr7_-_135083906
|
0.233
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr15_+_46285789
|
0.232
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr15_-_48434896
|
0.232
|
NM_002041
NM_005254
NM_016654
NM_016655
NM_181427
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1
|
|
chr1_+_67923331
|
0.230
|
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr6_-_127882192
|
0.230
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr11_+_34599227
|
0.230
|
|
EHF
|
ets homologous factor
|
|
chr5_+_140698537
|
0.229
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr4_-_75066533
|
0.229
|
NM_002619
|
PF4
|
platelet factor 4
|
|
chr21_-_35343331
|
0.229
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_-_102517196
|
0.229
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr10_+_68355797
|
0.227
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr6_+_130381408
|
0.226
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr15_+_48434397
|
0.225
|
|
LOC100129387
|
hypothetical LOC100129387
|
|
chr17_-_15495534
|
0.225
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr3_-_57653634
|
0.225
|
NM_152678
|
FAM116A
|
family with sequence similarity 116, member A
|
|
chr11_-_14337167
|
0.224
|
NM_012250
|
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2
|
|
chr5_-_58918031
|
0.223
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_10362583
|
0.223
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr18_-_29273979
|
0.223
|
NM_001105528
|
C18orf34
|
chromosome 18 open reading frame 34
|
|
chr14_-_75517844
|
0.222
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr2_+_28892366
|
0.222
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr11_+_34599163
|
0.222
|
NM_012153
|
EHF
|
ets homologous factor
|
|
chr10_+_70172257
|
0.221
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr8_-_30789865
|
0.221
|
NM_001009552
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr6_+_12825022
|
0.219
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr3_+_110338250
|
0.219
|
|
FLJ22763
|
hypothetical LOC401081
|
|
chr18_+_28023984
|
0.218
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr12_-_101398470
|
0.218
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_191422346
|
0.218
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr3_-_113841200
|
0.217
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_69736600
|
0.217
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr1_+_81544432
|
0.217
|
|
LPHN2
|
latrophilin 2
|
|
chr12_-_101398429
|
0.216
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_16217871
|
0.215
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr5_+_140509803
|
0.215
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr4_-_69116839
|
0.215
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr6_+_12824873
|
0.215
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chrX_-_77801480
|
0.214
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr8_+_98856984
|
0.213
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr13_-_29322671
|
0.212
|
|
UBL3
|
ubiquitin-like 3
|
|
chr6_-_76838944
|
0.212
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr4_+_169789327
|
0.212
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr5_+_161206774
|
0.211
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr21_+_38550739
|
0.211
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr3_+_162697289
|
0.210
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr4_-_176970895
|
0.209
|
|
GPM6A
|
glycoprotein M6A
|
|
chrX_-_15243647
|
0.208
|
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr6_-_28429950
|
0.208
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr19_-_3936343
|
0.208
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr2_-_200030412
|
0.207
|
|
SATB2
|
SATB homeobox 2
|
|
chr3_-_143230124
|
0.207
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr4_+_41395493
|
0.206
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_155818728
|
0.206
|
NM_015718
|
NOX3
|
NADPH oxidase 3
|
|
chr6_+_46869052
|
0.205
|
NM_005588
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase)
|
|
chr9_-_27287106
|
0.205
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr5_-_160044844
|
0.204
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chrX_+_1670485
|
0.204
|
NM_005088
|
AKAP17A
|
A kinase (PRKA) anchor protein 17A
|