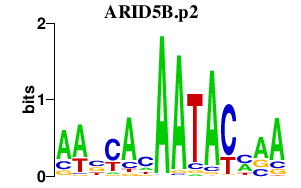
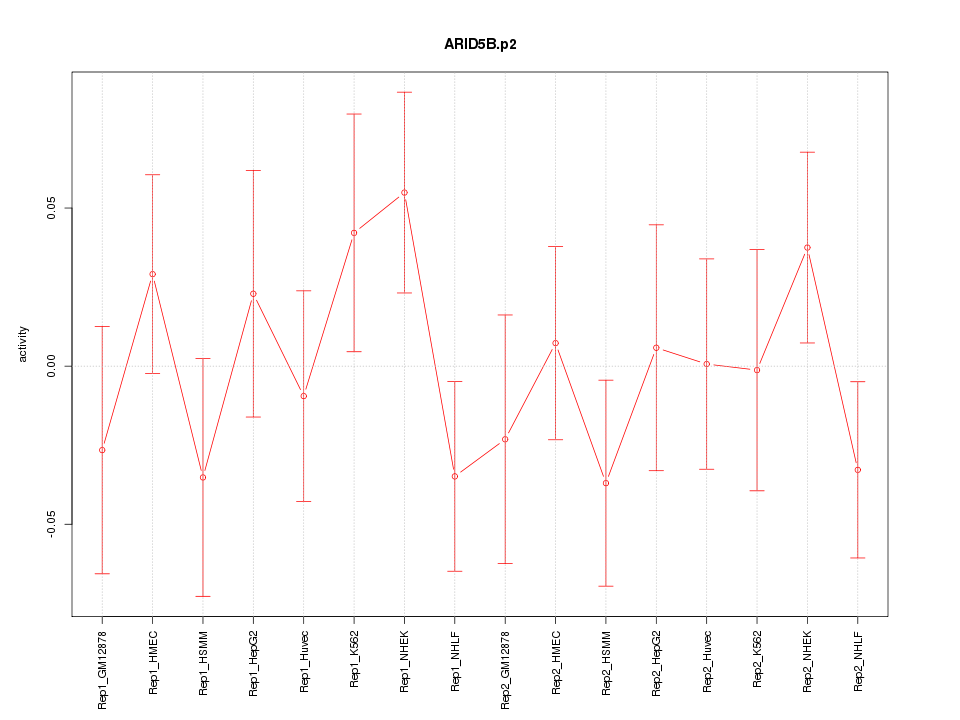
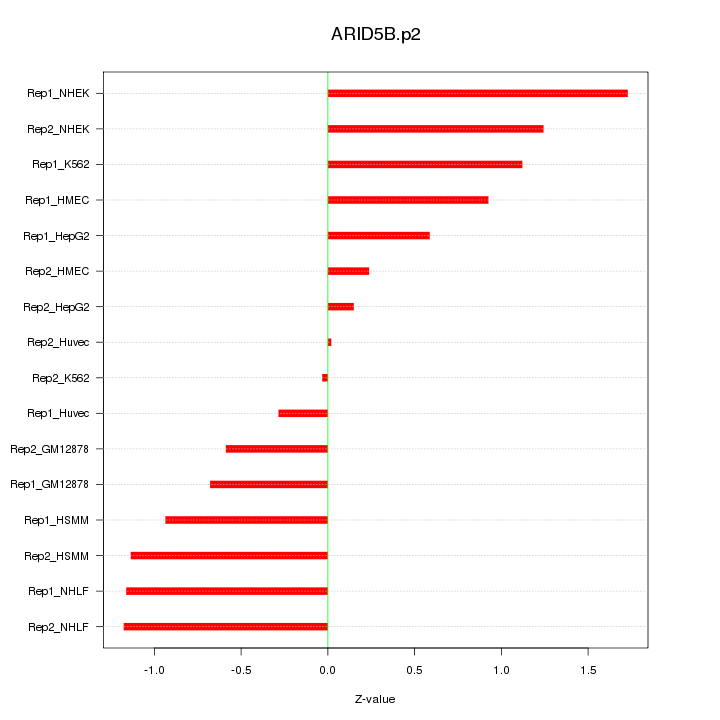
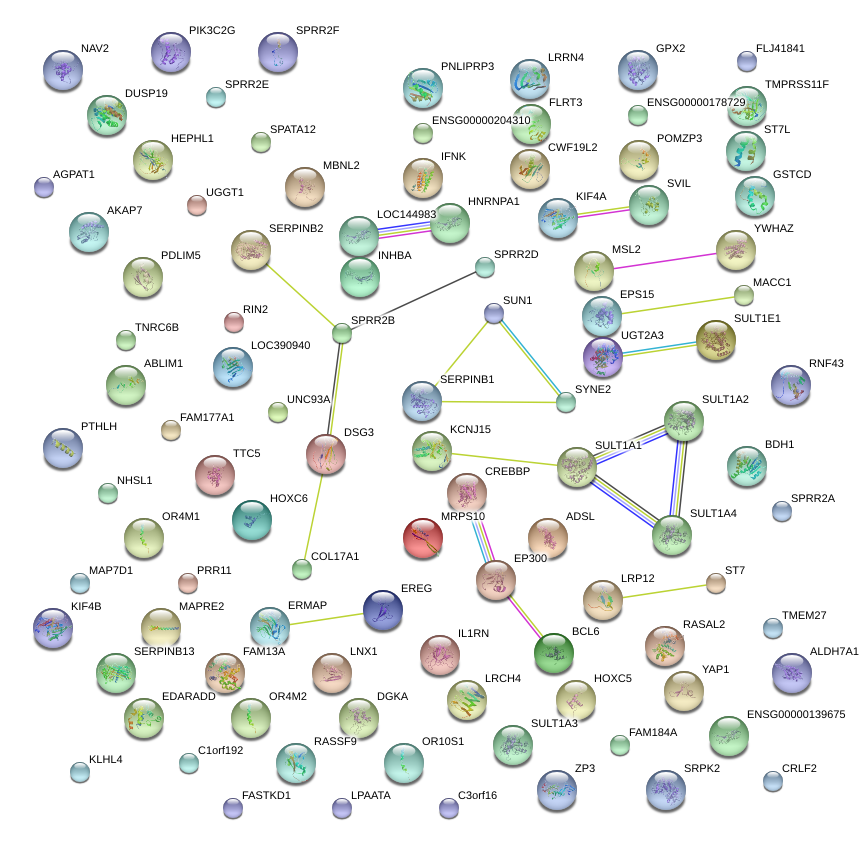
|
chr7_+_116447527
|
2.357
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr17_-_74244539
|
1.382
|
|
|
|
|
chr7_-_104696678
|
1.367
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr10_+_102113410
|
1.298
|
|
|
|
|
chr20_+_19863716
|
1.288
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr10_-_29963906
|
1.200
|
NM_021738
|
SVIL
|
supervillin
|
|
chr7_-_20223467
|
1.098
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr14_-_64479198
|
1.097
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr16_-_3783475
|
1.087
|
|
CREBBP
|
CREB binding protein
|
|
chr10_-_29963847
|
1.051
|
|
|
|
|
chr10_+_118177371
|
0.959
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr3_-_188946168
|
0.935
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chrX_-_15593074
|
0.899
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr1_-_159604287
|
0.868
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr5_-_125956996
|
0.782
|
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr1_-_151280201
|
0.774
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr3_-_150993299
|
0.734
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr7_+_116447499
|
0.730
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr10_-_105835525
|
0.689
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr9_+_27514311
|
0.689
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr18_+_59405513
|
0.684
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr11_+_93394025
|
0.663
|
NM_001098672
|
HEPHL1
|
hephaestin-like 1
|
|
chr8_-_102029997
|
0.633
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr4_-_70760381
|
0.624
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr12_-_28016182
|
0.623
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr22_+_38903825
|
0.573
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr6_-_48186868
|
0.569
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr20_-_14266200
|
0.560
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chrX_+_86659370
|
0.552
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr17_+_7682355
|
0.546
|
|
|
|
|
chr16_+_30118036
|
0.520
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr11_-_123353607
|
0.519
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr15_+_19869841
|
0.499
|
NM_001004719
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2
|
|
chr1_+_43063817
|
0.498
|
NM_018538
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chr16_+_29378694
|
0.495
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr2_+_113591940
|
0.461
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr4_+_106851415
|
0.455
|
NM_001031720
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr19_+_48772791
|
0.455
|
NM_001193621
|
LOC390940
|
hypothetical protein LOC390940
|
|
chr4_-_90197140
|
0.447
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr12_+_18305740
|
0.428
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr3_+_57069508
|
0.427
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr2_-_170138558
|
0.422
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr1_+_208067890
|
0.416
|
NM_014388
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr12_+_54611212
|
0.411
|
NM_001345
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr14_-_19843918
|
0.408
|
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chr2_+_183651756
|
0.403
|
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr10_-_105835609
|
0.402
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr7_+_75892187
|
0.399
|
NM_001110354
|
ZP3
|
zona pellucida glycoprotein 3 (sperm receptor)
|
|
chr4_+_75449720
|
0.386
|
NM_001432
|
EREG
|
epiregulin
|
|
chr11_+_101488380
|
0.370
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr1_+_208067974
|
0.368
|
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr6_-_138862273
|
0.366
|
|
|
|
|
chr1_+_208067986
|
0.357
|
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr11_-_106833715
|
0.355
|
NM_152434
|
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe)
|
|
chr8_-_11748330
|
0.355
|
|
|
|
|
chr17_+_54587882
|
0.347
|
|
PRR11
|
proline rich 11
|
|
chr2_+_128644510
|
0.347
|
|
UGGT1
|
UDP-glucose glycoprotein glucosyltransferase 1
|
|
chr12_-_84754289
|
0.345
|
NM_005447
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr7_+_838663
|
0.345
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr6_-_42293581
|
0.342
|
NM_018141
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr4_-_68678181
|
0.341
|
NM_207407
|
TMPRSS11F
|
transmembrane protease, serine 11F
|
|
chrX_-_1291526
|
0.338
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr13_+_96726197
|
0.323
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr11_+_20005704
|
0.322
|
|
NAV2
|
neuron navigator 2
|
|
chr6_-_2785682
|
0.318
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr12_+_52697160
|
0.314
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr18_+_27281729
|
0.307
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr10_-_116276651
|
0.305
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_-_53010929
|
0.303
|
|
|
|
|
chr14_+_63750611
|
0.302
|
NM_182913
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2
|
|
chr21_+_38550533
|
0.297
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr1_+_36409277
|
0.297
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr1_-_151310707
|
0.295
|
NM_001017418
|
SPRR2B
|
small proline-rich protein 2B
|
|
chr17_-_53847937
|
0.287
|
|
RNF43
|
ring finger protein 43
|
|
chr6_-_119512035
|
0.287
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr13_+_52114565
|
0.285
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr6_-_138862271
|
0.283
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr18_+_59705918
|
0.281
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr3_-_198784590
|
0.278
|
NM_203315
|
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr4_-_69852090
|
0.276
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr1_+_176577228
|
0.275
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr7_-_41709191
|
0.275
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr5_+_154373452
|
0.274
|
NM_001099293
|
KIF4B
|
kinesin family member 4B
|
|
chr20_-_5982617
|
0.273
|
NM_152611
|
LRRN4
|
leucine rich repeat neuronal 4
|
|
chr18_+_30810889
|
0.269
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr6_-_32247134
|
0.269
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr1_+_234624266
|
0.269
|
NM_145861
|
EDARADD
|
EDAR-associated death domain
|
|
chr3_-_137395966
|
0.266
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr6_+_167624792
|
0.254
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr11_+_43622348
|
0.253
|
|
|
|
|
chr4_-_151993422
|
0.252
|
|
|
|
|
chr4_+_95595419
|
0.248
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr1_-_51660328
|
0.244
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr1_-_151352587
|
0.244
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr1_-_151333624
|
0.242
|
NM_001024209
|
SPRR2E
|
small proline-rich protein 2E
|
|
chr14_+_34583863
|
0.241
|
NM_001079519
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr4_-_54152480
|
0.240
|
NM_001126328
|
LNX1
|
ligand of numb-protein X 1
|
|
chr1_+_556968
|
0.239
|
|
|
|
|
chr6_+_131508153
|
0.239
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr11_-_118405288
|
0.239
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr11_-_33139449
|
0.237
|
NM_001033505
NM_001033506
NM_001326
|
CSTF3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa
|
|
chr2_+_201806410
|
0.234
|
NM_001080124
NM_001228
NM_033355
NM_033358
|
CASP8
|
caspase 8, apoptosis-related cysteine peptidase
|
|
chr18_-_59480082
|
0.233
|
NM_006919
|
SERPINB3
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chr18_+_53865455
|
0.233
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_-_36594652
|
0.226
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr17_-_24265108
|
0.224
|
|
PHF12
|
PHD finger protein 12
|
|
chr1_+_234748187
|
0.219
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr1_+_208068022
|
0.218
|
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr7_+_133862883
|
0.218
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chr1_+_13784553
|
0.215
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr5_-_98290117
|
0.215
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr11_+_48466844
|
0.215
|
NM_001005512
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47
|
|
chr6_+_30238961
|
0.212
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr1_+_150005754
|
0.210
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr17_-_61655915
|
0.207
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr4_-_159313613
|
0.206
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr14_+_101346032
|
0.204
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr10_+_124135574
|
0.202
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr13_+_104916216
|
0.202
|
NM_001161814
NM_172370
|
DAOA
|
D-amino acid oxidase activator
|
|
chr3_+_11242666
|
0.201
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr3_-_52688768
|
0.201
|
NM_018165
NM_181042
|
PBRM1
|
polybromo 1
|
|
chr12_-_23995109
|
0.199
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_26550243
|
0.199
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr9_+_33740463
|
0.197
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|
|
chr1_+_557269
|
0.197
|
|
|
|
|
chr18_-_44723116
|
0.193
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr1_+_58899
|
0.193
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr2_-_99238001
|
0.192
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chrX_+_99726445
|
0.190
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr7_-_22518200
|
0.189
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr12_+_4628521
|
0.189
|
NM_005002
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa
|
|
chrX_-_33267646
|
0.188
|
NM_000109
|
DMD
|
dystrophin
|
|
chr6_+_71192862
|
0.186
|
NM_001162529
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr2_+_10101081
|
0.185
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr12_+_50633430
|
0.184
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr3_+_113287864
|
0.182
|
NM_001171747
NM_024616
|
C3orf52
|
chromosome 3 open reading frame 52
|
|
chr12_+_61645361
|
0.179
|
|
RPL14
|
ribosomal protein L14
|
|
chr1_-_203868622
|
0.179
|
NM_001973
NM_021795
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr15_+_47249700
|
0.175
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr17_-_78391178
|
0.175
|
NM_024702
|
ZNF750
|
zinc finger protein 750
|
|
chr2_-_74606819
|
0.174
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr9_-_116920232
|
0.173
|
NM_002160
|
TNC
|
tenascin C
|
|
chr20_+_31132978
|
0.173
|
NM_182519
|
C20orf186
|
chromosome 20 open reading frame 186
|
|
chr18_-_3209846
|
0.170
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr6_+_64344307
|
0.169
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr18_+_53863865
|
0.169
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr18_-_59462469
|
0.168
|
NM_002974
|
SERPINB4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 4
|
|
chrX_-_131375217
|
0.167
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr4_+_170817787
|
0.167
|
|
CLCN3
|
chloride channel 3
|
|
chr20_-_34921689
|
0.166
|
NM_199181
|
KIAA0889
|
KIAA0889
|
|
chrX_-_140614227
|
0.160
|
NM_145665
NM_032417
|
SPANXE
SPANXD
SPANXA2
|
SPANX family, member E
SPANX family, member D
SPANX family, member A2
|
|
chrX_-_105169370
|
0.158
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chrX_-_10761729
|
0.156
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_88208650
|
0.155
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr10_-_25344871
|
0.155
|
NM_145010
|
ENKUR
|
enkurin, TRPC channel interacting protein
|
|
chr14_-_19843969
|
0.154
|
NM_138376
|
TTC5
|
tetratricopeptide repeat domain 5
|
|
chrX_-_15263555
|
0.151
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr2_-_165133239
|
0.151
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr6_+_80397690
|
0.149
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr20_-_9767406
|
0.148
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr12_-_8706533
|
0.148
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr12_-_52868855
|
0.145
|
NM_014311
|
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1
|
|
chr1_-_42549347
|
0.143
|
|
|
|
|
chr6_+_64289585
|
0.143
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr1_-_148936123
|
0.141
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr7_-_33047042
|
0.140
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr3_+_142175329
|
0.139
|
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr1_+_51855343
|
0.136
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr12_-_49076658
|
0.136
|
NM_001145475
|
FAM186A
|
family with sequence similarity 186, member A
|
|
chr18_-_28247186
|
0.133
|
|
|
|
|
chr16_+_2068186
|
0.132
|
|
TSC2
|
tuberous sclerosis 2
|
|
chr7_-_96471544
|
0.132
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr14_+_101345878
|
0.132
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr14_+_69307914
|
0.132
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr2_+_119416214
|
0.131
|
NM_006770
|
MARCO
|
macrophage receptor with collagenous structure
|
|
chr14_+_38653177
|
0.130
|
NM_001009182
NM_001009183
NM_003616
|
SIP1
|
survival of motor neuron protein interacting protein 1
|
|
chr12_+_49522967
|
0.130
|
NM_182559
|
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12
|
|
chr3_+_43303005
|
0.129
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr13_+_48582389
|
0.128
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chrX_-_129864887
|
0.127
|
NM_006375
NM_182314
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2
|
|
chrX_-_48976714
|
0.127
|
NM_005183
|
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit
|
|
chr6_+_126702914
|
0.127
|
NM_001012507
|
CENPW
|
centromere protein W
|
|
chr2_+_170370290
|
0.127
|
|
SSB
|
Sjogren syndrome antigen B (autoantigen La)
|
|
chr6_-_86407875
|
0.127
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr10_-_64698831
|
0.126
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr19_-_53450953
|
0.125
|
NM_001184902
NM_001184904
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr3_+_138132006
|
0.124
|
NM_001190796
|
NCK1
|
NCK adaptor protein 1
|
|
chr5_+_36644187
|
0.124
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_-_48902692
|
0.124
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr12_-_8706637
|
0.123
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr15_-_53996597
|
0.122
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr18_+_18767275
|
0.121
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr3_-_180452338
|
0.121
|
NM_171830
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr1_+_76024466
|
0.120
|
NM_004582
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit
|
|
chr12_-_101036407
|
0.120
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr3_+_157877146
|
0.119
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr10_-_123343320
|
0.119
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_+_168383427
|
0.119
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chrX_+_30158473
|
0.118
|
NM_002365
|
MAGEB3
|
melanoma antigen family B, 3
|
|
chr14_-_22693333
|
0.118
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|