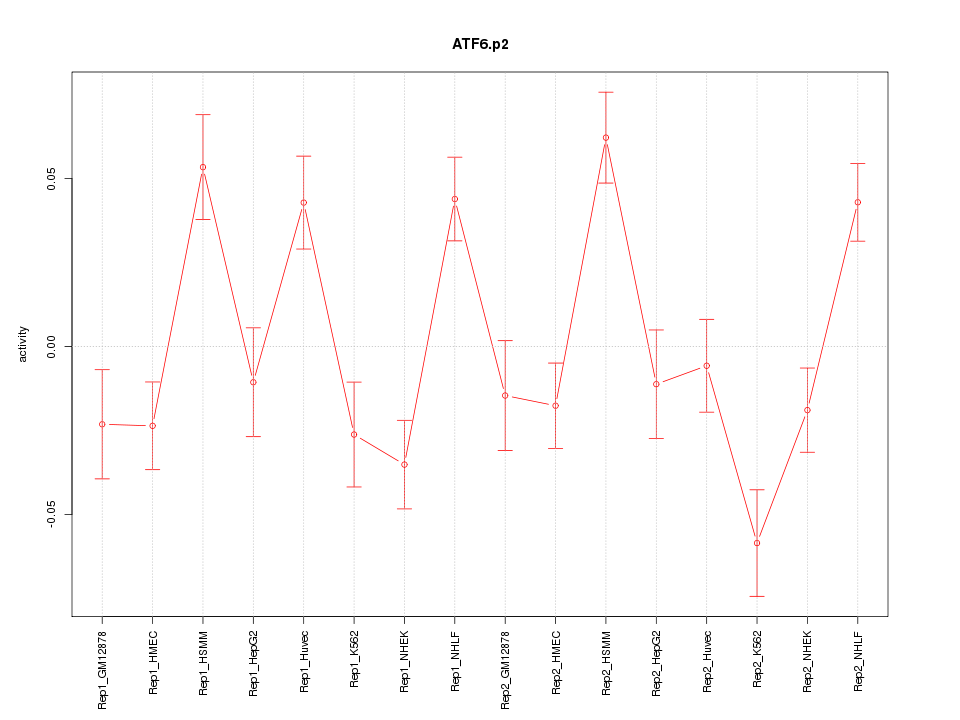
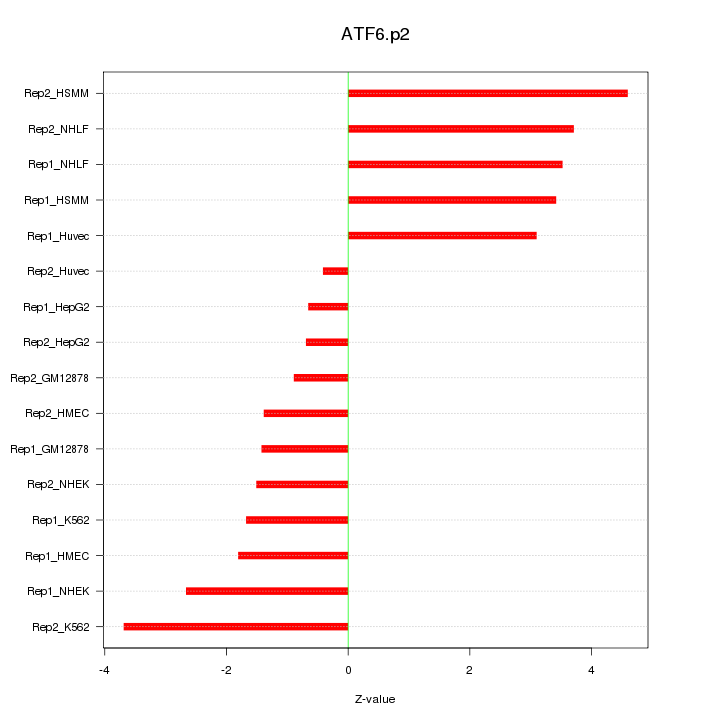
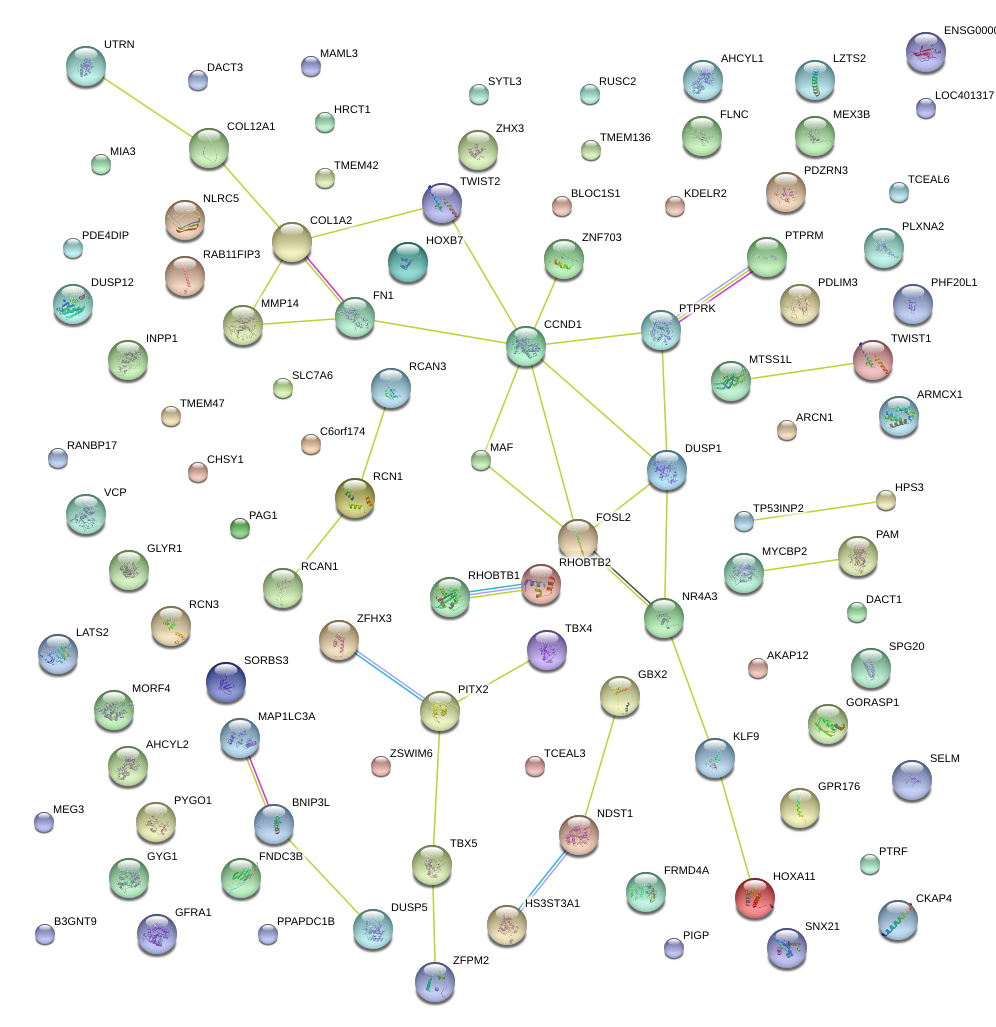
|
chr13_-_20533629
|
4.934
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_+_54723044
|
3.882
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr2_+_28469275
|
3.843
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_27191534
|
3.755
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr2_+_28469199
|
3.642
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr2_+_28469172
|
3.389
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr16_-_71639670
|
3.034
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_+_26296439
|
2.921
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr13_-_35818777
|
2.911
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr4_-_186693574
|
2.844
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr10_-_62373828
|
2.740
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr13_-_35818603
|
2.687
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr9_+_35528890
|
2.680
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr6_+_144649142
|
2.674
|
|
UTRN
|
utrophin
|
|
chr17_-_13445942
|
2.617
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr13_-_20533703
|
2.584
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chrX_+_100692071
|
2.578
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr9_+_35896188
|
2.524
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr7_-_27191287
|
2.503
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr13_-_35818491
|
2.475
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr17_-_44043361
|
2.445
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr9_+_35528628
|
2.422
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr12_-_123568781
|
2.402
|
|
|
|
|
chr13_-_35818396
|
2.350
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr3_-_73756668
|
2.347
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr3_+_44878380
|
2.286
|
NM_144638
|
TMEM42
|
transmembrane protein 42
|
|
chr17_-_44043278
|
2.278
|
|
HOXB7
|
homeobox B7
|
|
chr8_-_38245694
|
2.273
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr16_+_415606
|
2.247
|
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr8_+_106400322
|
2.202
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr11_+_119701047
|
2.185
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr16_-_65742323
|
2.117
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr6_-_128883125
|
2.073
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr5_-_172130766
|
2.060
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr12_+_54396099
|
2.046
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr8_-_82186832
|
2.033
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr1_+_220857774
|
2.003
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr1_+_110328850
|
1.985
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chrX_-_34585287
|
1.975
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_+_150330060
|
1.963
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr12_+_54396118
|
1.927
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr5_+_102229425
|
1.925
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr12_+_54396086
|
1.890
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr1_+_110328922
|
1.888
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr15_-_38000379
|
1.878
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr10_+_102749597
|
1.851
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr7_-_6490375
|
1.846
|
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr9_+_101623957
|
1.842
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr12_-_105165805
|
1.837
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr7_-_6490214
|
1.835
|
NM_001100603
NM_006854
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr7_+_93861808
|
1.830
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr4_-_187884721
|
1.821
|
|
|
|
|
chr3_-_131312963
|
1.779
|
|
|
|
|
chr15_-_80125514
|
1.777
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr15_-_80125403
|
1.777
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr7_+_28415495
|
1.773
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr7_-_19123787
|
1.764
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr18_+_7557313
|
1.742
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr7_+_128257698
|
1.737
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr14_+_22375753
|
1.735
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr7_-_19123765
|
1.726
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr16_+_55580910
|
1.724
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr5_+_102229612
|
1.704
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_+_149845503
|
1.701
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr3_-_75566781
|
1.688
|
|
FAM86DP
|
family with sequence similarity 86, member D, pseudogene
|
|
chr15_-_99609197
|
1.687
|
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr3_+_173240126
|
1.667
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr20_+_32610161
|
1.658
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr9_-_72218098
|
1.653
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr1_-_206484258
|
1.633
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr3_-_8518327
|
1.623
|
|
LOC100288428
|
hypothetical LOC100288428
|
|
chr4_-_141293572
|
1.622
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr14_+_22375632
|
1.606
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr6_-_127822227
|
1.585
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr14_+_22375747
|
1.575
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr1_+_110328830
|
1.571
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr11_+_119701214
|
1.571
|
|
TMEM136
|
transmembrane protein 136
|
|
chr17_+_56884560
|
1.569
|
|
TBX4
|
T-box 4
|
|
chr16_+_66855827
|
1.558
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6
|
|
chr17_-_44042924
|
1.548
|
|
HOXB7
|
homeobox B7
|
|
chr4_-_111763655
|
1.541
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr3_-_131313010
|
1.537
|
|
|
|
|
chr4_-_111763356
|
1.531
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr15_-_53668341
|
1.531
|
NM_015617
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr14_+_22375656
|
1.526
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr20_-_39379533
|
1.520
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr10_+_112247614
|
1.517
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr3_+_173240107
|
1.511
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr16_-_78192111
|
1.505
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr20_+_32755778
|
1.503
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr1_+_110328941
|
1.497
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr14_+_58174489
|
1.486
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr6_+_151602787
|
1.479
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_+_60663842
|
1.457
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr17_-_37828643
|
1.456
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr12_-_113328271
|
1.453
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr8_+_133856738
|
1.450
|
NM_016018
NM_032205
NM_198513
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr5_+_170221584
|
1.444
|
NM_022897
|
RANBP17
|
RAN binding protein 17
|
|
chr11_+_69165119
|
1.443
|
|
CCND1
|
cyclin D1
|
|
chr8_+_22479115
|
1.427
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr8_+_37672427
|
1.427
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr10_-_118022965
|
1.425
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr22_-_29833484
|
1.412
|
NM_080430
|
SELM
|
selenoprotein M
|
|
chr5_-_99898788
|
1.399
|
|
|
|
|
chr6_-_75972473
|
1.397
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr3_+_127118130
|
1.394
|
|
LOC100125556
|
family with sequence similarity 86, member A pseudogene
|
|
chr15_+_76952356
|
1.394
|
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr14_+_100362204
|
1.394
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr20_+_43895802
|
1.374
|
NM_001042632
NM_001042633
NM_033421
NM_152897
|
SNX21
|
sorting nexin family member 21
|
|
chrX_+_102749489
|
1.373
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr16_-_4837266
|
1.372
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr2_-_236741352
|
1.329
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr8_+_133856793
|
1.326
|
|
PHF20L1
|
PHD finger protein 20-like 1
|
|
chr11_+_69165065
|
1.317
|
|
CCND1
|
cyclin D1
|
|
chr2_-_216008689
|
1.317
|
|
FN1
|
fibronectin 1
|
|
chr15_-_99609648
|
1.313
|
NM_014918
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr21_-_37366684
|
1.309
|
NM_153681
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr2_-_216009015
|
1.303
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr3_+_150191884
|
1.280
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr21_-_37367259
|
1.278
|
NM_153682
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr11_+_117948350
|
1.273
|
|
ARCN1
|
archain 1
|
|
chr16_-_69277355
|
1.269
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr9_-_35062643
|
1.267
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr11_+_117948301
|
1.265
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr3_+_173240136
|
1.260
|
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_-_143750988
|
1.259
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr10_-_14090459
|
1.239
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr6_+_158990982
|
1.233
|
NM_001009991
|
SYTL3
|
synaptotagmin-like 3
|
|
chr11_+_32068990
|
1.232
|
NM_002901
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr2_+_190916571
|
1.231
|
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr11_+_69165018
|
1.226
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr3_-_39124124
|
1.224
|
NM_031899
|
GORASP1
|
golgi reassembly stacking protein 1, 65kDa
|
|
chr3_-_49370682
|
1.218
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr14_+_23905984
|
1.215
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr9_-_35062545
|
1.212
|
|
VCP
|
valosin containing protein
|
|
chr3_-_109423858
|
1.207
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr19_-_18493886
|
1.207
|
NM_006532
|
ELL
|
elongation factor RNA polymerase II
|
|
chr12_+_58276114
|
1.196
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr9_-_35062596
|
1.194
|
|
VCP
|
valosin containing protein
|
|
chr14_-_34253473
|
1.184
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_-_77824849
|
1.183
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr3_-_49370755
|
1.180
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr3_+_39124155
|
1.164
|
NM_001105513
NM_145755
|
TTC21A
|
tetratricopeptide repeat domain 21A
|
|
chr12_+_120635021
|
1.163
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr7_-_130443379
|
1.133
|
|
FLJ43663
|
hypothetical LOC378805
|
|
chr7_-_37922902
|
1.126
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr22_+_34107087
|
1.124
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr9_-_35062578
|
1.118
|
|
VCP
|
valosin containing protein
|
|
chr12_+_970613
|
1.114
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr21_+_37367432
|
1.102
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr14_+_100362236
|
1.096
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_+_69165159
|
1.094
|
|
CCND1
|
cyclin D1
|
|
chr1_-_158091296
|
1.079
|
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr3_-_39124036
|
1.075
|
|
GORASP1
|
golgi reassembly stacking protein 1, 65kDa
|
|
chr7_-_129379947
|
1.065
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr22_+_34107051
|
1.064
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr7_-_37923040
|
1.058
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr18_+_54682261
|
1.056
|
|
ZNF532
|
zinc finger protein 532
|
|
chr2_-_1727292
|
1.055
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr6_+_1336077
|
1.053
|
|
|
|
|
chr17_+_77529070
|
1.049
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr13_-_44048391
|
1.035
|
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr16_-_65775326
|
1.020
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr21_-_43671355
|
1.016
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr19_+_52451570
|
1.014
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr14_+_92869317
|
1.013
|
NM_020818
|
KIAA1409
|
KIAA1409
|
|
chr5_+_170221441
|
1.008
|
|
RANBP17
|
RAN binding protein 17
|
|
chr19_+_52451642
|
1.001
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr3_+_101694098
|
1.001
|
NM_018004
|
TMEM45A
|
transmembrane protein 45A
|
|
chr10_-_98336548
|
0.997
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr1_-_59020875
|
0.994
|
|
JUN
|
jun proto-oncogene
|
|
chr7_+_128166581
|
0.988
|
NM_001130674
NM_001219
|
CALU
|
calumenin
|
|
chr2_-_198248828
|
0.985
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr6_-_75972286
|
0.985
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr22_+_37193961
|
0.984
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr9_-_74169649
|
0.984
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr19_-_45888259
|
0.983
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr20_-_2769245
|
0.983
|
NM_022760
|
FAM113A
|
family with sequence similarity 113, member A
|
|
chr12_+_20413445
|
0.979
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr6_-_86409701
|
0.973
|
NM_001159677
NM_006372
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_-_33864863
|
0.971
|
|
LEMD2
|
LEM domain containing 2
|
|
chr19_-_18493847
|
0.970
|
|
ELL
|
elongation factor RNA polymerase II
|
|
chr7_+_30140868
|
0.967
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr16_+_2510323
|
0.966
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr6_-_90119068
|
0.961
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr10_+_99486881
|
0.961
|
|
ZFYVE27
|
zinc finger, FYVE domain containing 27
|
|
chr12_-_51759400
|
0.960
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr4_-_119976736
|
0.959
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr4_-_119976631
|
0.957
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr7_+_150560459
|
0.957
|
NM_019015
|
CHPF2
|
chondroitin polymerizing factor 2
|
|
chr16_+_15435825
|
0.956
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr11_+_101291134
|
0.954
|
|
KIAA1377
|
KIAA1377
|
|
chr18_+_9903954
|
0.954
|
NM_003574
NM_194434
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr5_+_53849251
|
0.952
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chr15_-_53667800
|
0.951
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr10_-_131652364
|
0.945
|
|
EBF3
|
early B-cell factor 3
|
|
chr12_+_70434927
|
0.945
|
|
RAB21
|
RAB21, member RAS oncogene family
|
|
chr16_-_5087740
|
0.937
|
NM_201400
NM_201598
|
FAM86A
|
family with sequence similarity 86, member A
|
|
chr6_-_90119158
|
0.937
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr4_-_114901789
|
0.927
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|