|
chr19_+_58750305
|
2.302
|
NM_001079907
|
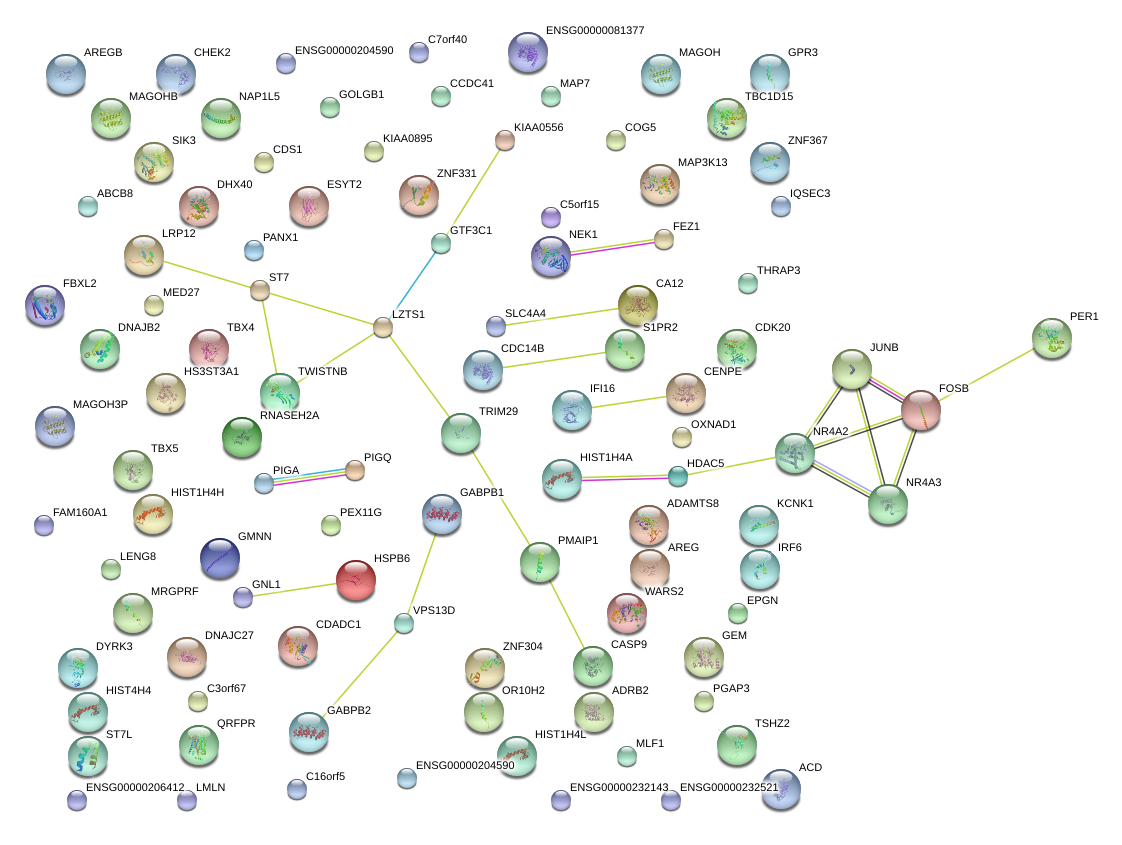
ZNF331
|
zinc finger protein 331
|
|
chr2_-_156897286
|
2.004
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr5_+_148186348
|
1.563
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr12_-_113328271
|
1.480
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr4_+_75529716
|
1.458
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr19_+_50663089
|
1.446
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr1_+_204875374
|
1.361
|
NM_001004023
NM_003582
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr11_-_119504726
|
1.313
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr19_-_10202947
|
1.301
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr20_+_51023159
|
1.282
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_+_119484529
|
1.265
|
|
|
|
|
chr12_-_10657397
|
1.251
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr5_-_133332173
|
1.241
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr4_-_187884721
|
1.218
|
|
|
|
|
chr14_-_49399317
|
1.215
|
|
RN7SL2
|
RNA, 7SL, cytoplasmic 2
|
|
chr1_-_119484733
|
1.178
|
NM_015836
NM_201263
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr11_-_116474196
|
1.166
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr1_+_204875671
|
1.136
|
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr4_+_75699652
|
1.116
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr4_+_152549776
|
1.089
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr17_+_56884560
|
1.079
|
|
TBX4
|
T-box 4
|
|
chr4_+_85723080
|
1.079
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr19_+_62554445
|
1.074
|
NM_020657
|
ZNF304
|
zinc finger protein 304
|
|
chr18_+_11841388
|
1.070
|
NM_020412
|
CHMP1B
|
chromatin modifying protein 1B
|
|
chr17_-_8000362
|
1.063
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr9_-_89779180
|
1.038
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr7_+_116447499
|
1.011
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr12_-_93377742
|
0.998
|
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr1_+_27591738
|
0.993
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr1_+_231816372
|
0.968
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr1_-_208046011
|
0.957
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr7_-_19715127
|
0.957
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr17_-_13445942
|
0.946
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr7_-_158314963
|
0.945
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr11_-_68537243
|
0.934
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr11_-_124871258
|
0.911
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr7_-_36372971
|
0.911
|
NM_015314
|
KIAA0895
|
KIAA0895
|
|
chr8_-_95343716
|
0.909
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr18_+_55718171
|
0.903
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr7_+_150356435
|
0.889
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr16_-_66251611
|
0.882
|
|
ACD
|
adrenocortical dysplasia homolog (mouse)
|
|
chr12_-_51734434
|
0.877
|
|
LOC283335
|
hypothetical LOC283335
|
|
chr2_+_219852268
|
0.871
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr7_-_44992729
|
0.867
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr13_+_48720047
|
0.867
|
NM_001193478
NM_030911
|
CDADC1
|
cytidine and dCMP deaminase domain containing 1
|
|
chr19_-_40938251
|
0.866
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr8_-_95343696
|
0.857
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr12_-_93377584
|
0.845
|
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr3_+_199171467
|
0.842
|
NM_001136049
NM_033029
|
LMLN
|
leishmanolysin-like (metallopeptidase M8 family)
|
|
chr4_+_153676857
|
0.837
|
|
DKFZP434I0714
|
hypothetical protein DKFZP434I0714
|
|
chr3_+_186483408
|
0.835
|
|
LOC100505749
MAP3K13
|
hypothetical LOC100505749
mitogen-activated protein kinase kinase kinase 13
|
|
chr4_-_89837951
|
0.819
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr3_+_159771646
|
0.808
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr4_-_170770111
|
0.807
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr19_+_59652192
|
0.806
|
|
LENG8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr16_+_27468942
|
0.806
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr6_-_136913235
|
0.803
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr17_+_54997667
|
0.803
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr4_+_75393053
|
0.801
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr15_-_61461127
|
0.800
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr19_+_12763285
|
0.799
|
|
JUNB
|
jun B proto-oncogene
|
|
chr7_+_116447527
|
0.797
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr7_-_158314945
|
0.787
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr3_-_59010797
|
0.787
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr9_-_133945064
|
0.775
|
NM_004269
|
CRSP8P
MED27
|
mediator complex subunit 27 pseudogene
mediator complex subunit 27
|
|
chr9_+_101623957
|
0.774
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr3_-_59010777
|
0.770
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr3_+_33293915
|
0.768
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr3_-_59010740
|
0.759
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr12_+_46133
|
0.756
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr16_-_27468717
|
0.748
|
|
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa
|
|
chr3_+_16281670
|
0.748
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chrX_-_15263555
|
0.746
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr16_-_4528380
|
0.745
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr15_-_61460967
|
0.742
|
|
CA12
|
carbonic anhydrase XII
|
|
chrX_-_15263586
|
0.740
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr7_-_106991941
|
0.735
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chrX_-_15263596
|
0.734
|
NM_002641
NM_020473
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr4_+_72271218
|
0.733
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_-_104338924
|
0.727
|
NM_001813
|
CENPE
|
centromere protein E, 312kDa
|
|
chr9_-_98421894
|
0.725
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr12_+_70519753
|
0.724
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr13_+_44813479
|
0.722
|
|
LOC100190939
|
hypothetical LOC100190939
|
|
chr19_-_7459867
|
0.717
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr19_-_40938046
|
0.716
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr2_-_25047988
|
0.711
|
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr4_-_122521562
|
0.711
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr6_-_15356865
|
0.702
|
|
|
|
|
chr1_-_15723363
|
0.692
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr3_-_122951200
|
0.686
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr12_-_93377827
|
0.685
|
NM_001042399
NM_016122
|
CCDC41
|
coiled-coil domain containing 41
|
|
chr1_+_36462583
|
0.682
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr17_-_39556535
|
0.681
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr2_-_25048276
|
0.681
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr6_-_26393705
|
0.672
|
NM_003543
|
HIST1H4H
|
histone cluster 1, H4h
|
|
chr11_+_93501811
|
0.668
|
|
PANX1
|
pannexin 1
|
|
chr6_-_30631481
|
0.668
|
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr1_+_149309703
|
0.665
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr6_+_126702914
|
0.665
|
NM_001012507
|
CENPW
|
centromere protein W
|
|
chr9_-_133944877
|
0.663
|
|
MED27
|
mediator complex subunit 27
|
|
chr1_+_11963091
|
0.661
|
|
MFN2
|
mitofusin 2
|
|
chr10_+_101409159
|
0.659
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr11_+_93501791
|
0.653
|
|
PANX1
|
pannexin 1
|
|
chr11_-_10519265
|
0.653
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr5_+_108112537
|
0.647
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr4_-_170167962
|
0.647
|
NM_032783
|
CBR4
|
carbonyl reductase 4
|
|
chr8_+_142207866
|
0.645
|
NM_014957
|
DENND3
|
DENN/MADD domain containing 3
|
|
chr5_+_49998528
|
0.641
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr3_-_199171234
|
0.633
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr6_-_38715901
|
0.633
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr19_+_12763295
|
0.633
|
NM_002229
|
JUNB
|
jun B proto-oncogene
|
|
chr3_+_10003579
|
0.632
|
|
LOC442075
|
hypothetical LOC442075
|
|
chr4_-_120595218
|
0.632
|
|
|
|
|
chr3_+_95181649
|
0.630
|
NM_001174150
NM_001174151
NM_144996
NM_182896
|
ARL13B
|
ADP-ribosylation factor-like 13B
|
|
chr11_+_10519394
|
0.629
|
|
LOC100129827
|
hypothetical LOC100129827
|
|
chr9_-_168913
|
0.626
|
NM_001145355
NM_001145356
NM_018491
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr1_+_40399618
|
0.626
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr11_-_76862197
|
0.624
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr1_-_159413833
|
0.618
|
NM_003779
|
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3
|
|
chr9_+_70978790
|
0.618
|
NM_001170630
NM_004817
NM_201629
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr19_+_47593137
|
0.618
|
|
|
|
|
chr11_+_93501708
|
0.615
|
NM_015368
|
PANX1
|
pannexin 1
|
|
chr12_-_121946581
|
0.614
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr17_-_39556483
|
0.612
|
|
HDAC5
|
histone deacetylase 5
|
|
chr16_+_31036474
|
0.609
|
NM_032188
NM_182958
|
MYST1
|
MYST histone acetyltransferase 1
|
|
chr1_-_154147787
|
0.608
|
NM_006912
|
RIT1
|
Ras-like without CAAX 1
|
|
chr4_-_148824585
|
0.607
|
NM_138364
|
PRMT10
|
protein arginine methyltransferase 10 (putative)
|
|
chr6_-_38715567
|
0.607
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr6_+_35335456
|
0.606
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr7_-_93471463
|
0.605
|
NM_005868
|
BET1
|
blocked early in transport 1 homolog (S. cerevisiae)
|
|
chrX_+_16647582
|
0.605
|
NM_032796
|
SYAP1
|
synapse associated protein 1
|
|
chr3_+_115258270
|
0.604
|
NM_024638
|
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1
|
|
chr1_+_149247519
|
0.603
|
NM_021222
|
PRUNE
|
prune homolog (Drosophila)
|
|
chr7_+_28415495
|
0.600
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr9_-_109290528
|
0.599
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr15_-_27349300
|
0.597
|
NM_138704
|
NDNL2
|
necdin-like 2
|
|
chr3_-_38046034
|
0.594
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr3_-_16281287
|
0.593
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr17_-_46553083
|
0.592
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr2_+_192251105
|
0.590
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr19_+_62566690
|
0.590
|
NM_173631
|
TRAPPC2P1
ZNF547
|
trafficking protein particle complex 2 pseudogene 1
zinc finger protein 547
|
|
chr6_+_15357013
|
0.590
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr1_+_40399642
|
0.587
|
|
RLF
|
rearranged L-myc fusion
|
|
chr19_-_60320592
|
0.586
|
NM_017607
|
PPP1R12C
|
protein phosphatase 1, regulatory (inhibitor) subunit 12C
|
|
chr2_+_227408914
|
0.578
|
NM_001167608
NM_032276
|
RHBDD1
|
rhomboid domain containing 1
|
|
chr3_+_186786655
|
0.577
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr4_+_119732338
|
0.577
|
|
LOC729218
|
hypothetical LOC729218
|
|
chrX_+_16647653
|
0.573
|
|
SYAP1
|
synapse associated protein 1
|
|
chr5_+_95008236
|
0.572
|
NM_001131066
NM_001131065
NM_173362
|
RFESD
|
Rieske (Fe-S) domain containing
|
|
chr6_-_38715613
|
0.570
|
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr2_+_46623396
|
0.570
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr1_+_169721260
|
0.565
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr1_+_70649451
|
0.562
|
NM_001190463
NM_001902
NM_153742
|
CTH
|
cystathionase (cystathionine gamma-lyase)
|
|
chr14_+_69148066
|
0.561
|
|
KIAA0247
|
KIAA0247
|
|
chr1_+_32443822
|
0.560
|
NM_001160042
NM_018134
|
IQCC
|
IQ motif containing C
|
|
chr22_-_37062114
|
0.556
|
|
CSNK1E
|
casein kinase 1, epsilon
|
|
chr7_+_106991638
|
0.549
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr3_-_109423858
|
0.549
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr12_+_4253143
|
0.548
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr2_+_113911737
|
0.541
|
NM_172003
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr3_-_115258053
|
0.539
|
NM_020817
|
KIAA1407
|
KIAA1407
|
|
chr7_-_106991572
|
0.534
|
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr9_-_109291866
|
0.532
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr3_+_158026769
|
0.531
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr1_-_703848
|
0.530
|
|
LOC100288069
|
hypothetical LOC100288069
|
|
chr13_-_44813308
|
0.530
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr20_-_42584006
|
0.530
|
|
SERINC3
|
serine incorporator 3
|
|
chr12_+_22669352
|
0.527
|
|
ETNK1
|
ethanolamine kinase 1
|
|
chr18_+_18767275
|
0.523
|
NM_002894
|
RBBP8
|
retinoblastoma binding protein 8
|
|
chr6_+_116798791
|
0.523
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr1_+_11963062
|
0.522
|
|
MFN2
|
mitofusin 2
|
|
chr11_-_67029349
|
0.518
|
NM_001130848
NM_004910
|
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1
|
|
chr6_-_34772495
|
0.518
|
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr4_+_119419354
|
0.517
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr4_+_170778265
|
0.516
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr14_+_99017485
|
0.516
|
NM_001099402
|
CCNK
|
cyclin K
|
|
chr6_+_35334663
|
0.516
|
|
ZNF76
|
zinc finger protein 76
|
|
chr6_-_18230829
|
0.515
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr17_+_25467948
|
0.514
|
NM_032141
|
CCDC55
|
coiled-coil domain containing 55
|
|
chr21_-_42060266
|
0.513
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr6_-_108502547
|
0.513
|
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr4_-_186584031
|
0.512
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr1_-_149247464
|
0.512
|
NM_001163258
NM_018379
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr7_+_17304707
|
0.511
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr4_-_80079393
|
0.508
|
NM_001040202
|
PAQR3
|
progestin and adipoQ receptor family member III
|
|
chr17_+_44325127
|
0.506
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr11_+_34083784
|
0.505
|
|
NAT10
|
N-acetyltransferase 10 (GCN5-related)
|
|
chr3_-_130362563
|
0.505
|
NM_020701
|
ISY1
|
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr1_-_20685314
|
0.505
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr8_+_128875902
|
0.503
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr14_+_69148062
|
0.503
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr11_+_110978309
|
0.502
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr6_+_56927882
|
0.500
|
|
BEND6
|
BEN domain containing 6
|
|
chr16_-_47201588
|
0.497
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr3_-_158360576
|
0.493
|
|
CCNL1
|
cyclin L1
|
|
chr16_-_27468723
|
0.492
|
|
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa
|
|
chr6_+_35335278
|
0.489
|
|
ZNF76
|
zinc finger protein 76
|
|
chr17_+_18105148
|
0.486
|
NM_148886
|
SMCR7
|
Smith-Magenis syndrome chromosome region, candidate 7
|
|
chr3_-_64186151
|
0.485
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr9_+_96528814
|
0.483
|
|
C9orf3
|
chromosome 9 open reading frame 3
|