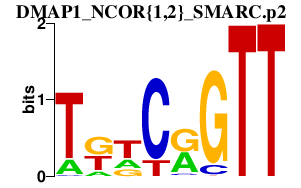
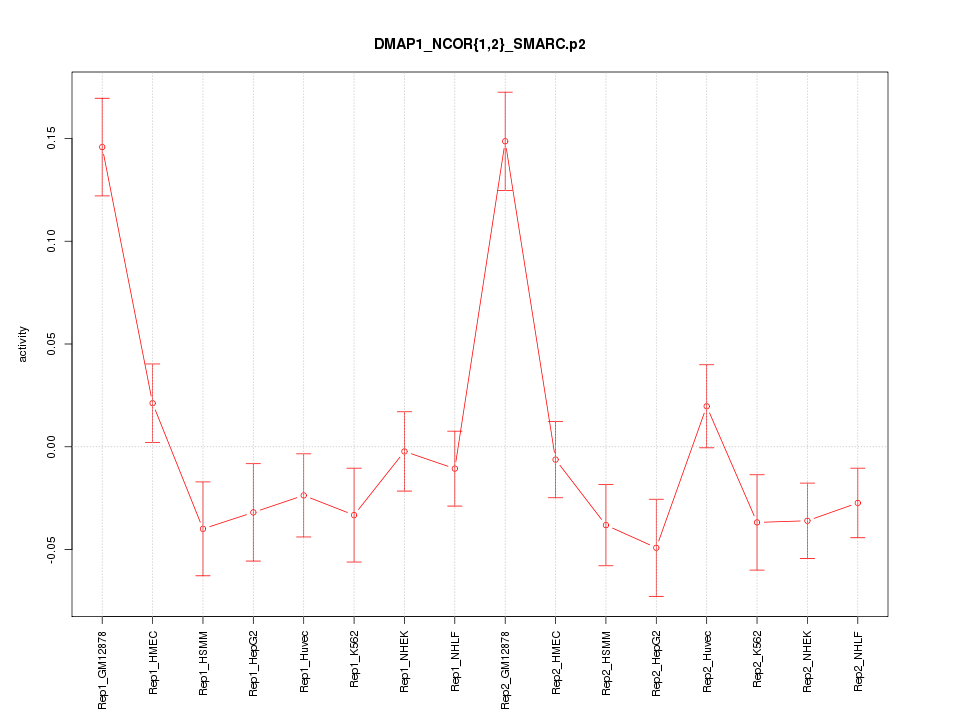
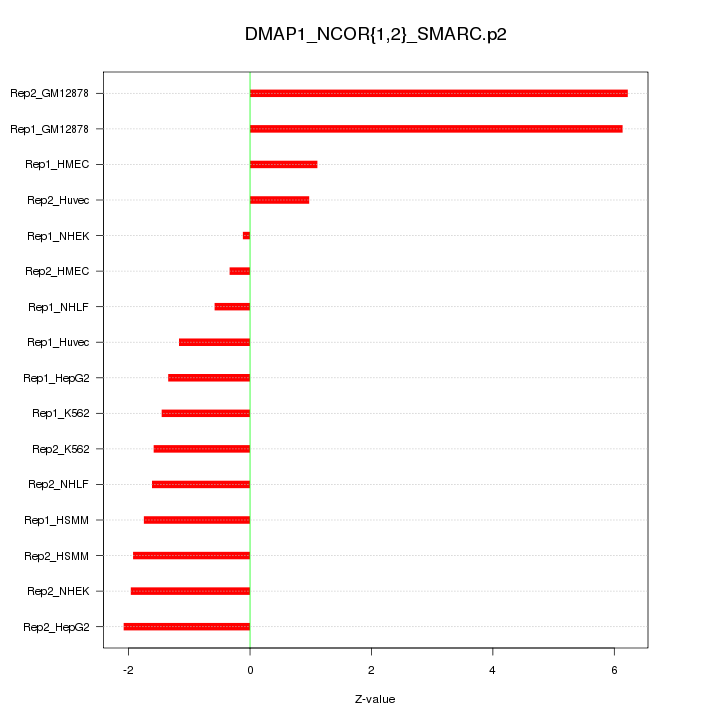
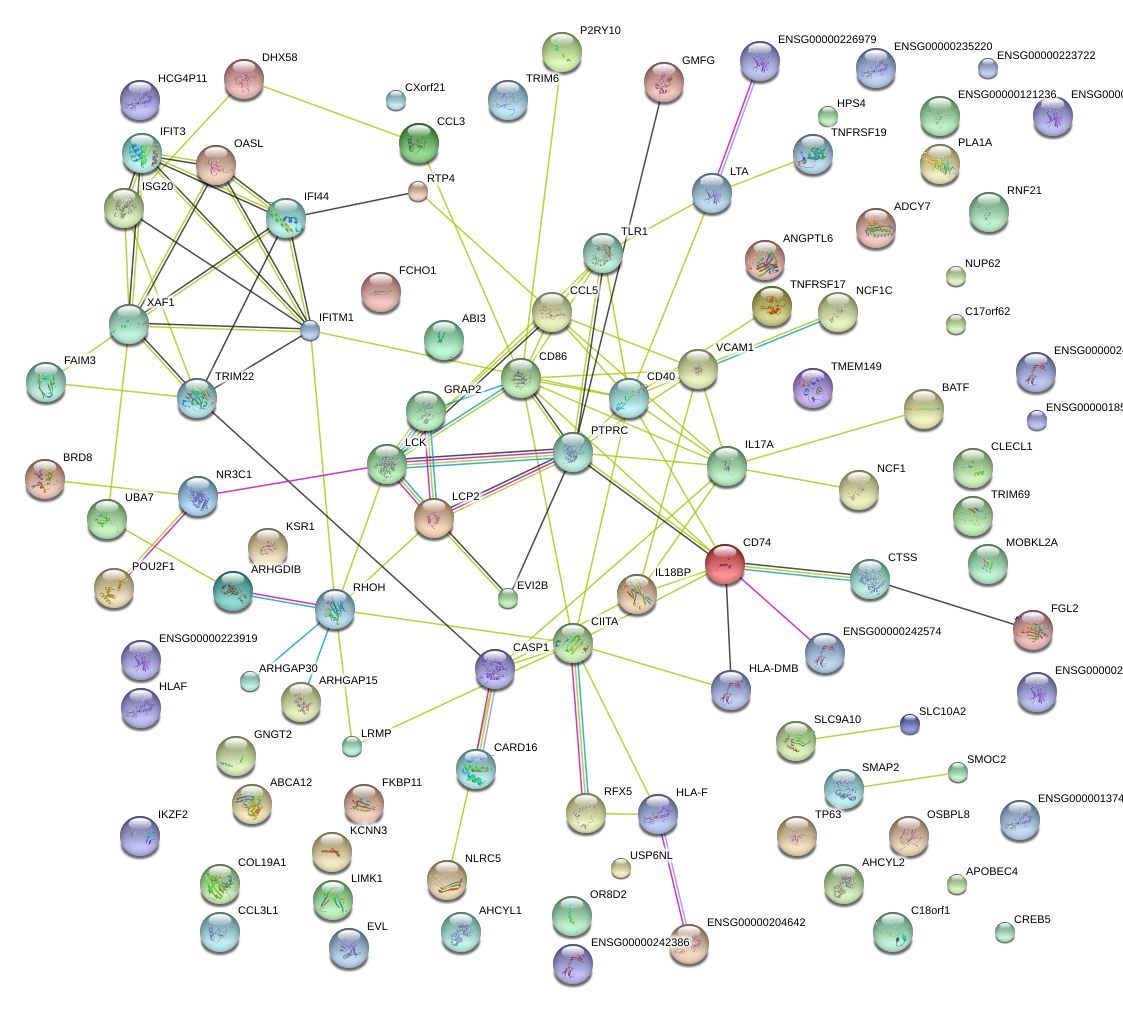
|
chr3_+_120799411
|
8.965
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr15_+_42815851
|
8.836
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr10_-_11614279
|
6.805
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chrX_+_78087484
|
6.667
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr5_-_138753443
|
6.400
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chrX_-_30505804
|
6.133
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr16_+_48865521
|
5.810
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr13_+_23042722
|
5.499
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr7_+_73826244
|
5.335
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr3_+_123279439
|
5.321
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr17_+_22823151
|
5.065
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr16_+_10878539
|
4.976
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr2_+_143603439
|
4.842
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr14_+_99601498
|
4.838
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr2_+_143603368
|
4.799
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr14_+_99601517
|
4.780
|
|
EVL
|
Enah/Vasp-like
|
|
chr1_+_165564904
|
4.719
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr19_-_10074332
|
4.508
|
NM_031917
|
ANGPTL6
|
angiopoietin-like 6
|
|
chr14_+_75058518
|
4.492
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr12_-_15005761
|
4.367
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr6_-_33016561
|
4.346
|
NM_002118
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta
|
|
chr1_-_205161821
|
4.339
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr17_-_78000890
|
4.305
|
NM_001193655
|
C17orf62
|
chromosome 17 open reading frame 62
|
|
chr14_+_75058591
|
4.299
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr1_+_165565006
|
4.090
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr11_+_5602782
|
3.922
|
NM_021616
|
TRIM34
|
tripartite motif containing 34
|
|
chr6_+_52159143
|
3.669
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chr17_+_6600080
|
3.577
|
|
XAF1
|
XIAP associated factor 1
|
|
chr1_+_32489426
|
3.408
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr3_-_113495763
|
3.359
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr3_+_188568861
|
3.304
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr5_-_142795269
|
3.265
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr5_-_149772528
|
3.224
|
|
|
|
|
chr1_-_149586298
|
3.174
|
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr20_+_44180296
|
3.123
|
NM_001250
NM_152854
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr5_-_149772469
|
3.123
|
NM_001025159
NM_004355
NM_001025158
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr20_+_44180355
|
3.117
|
|
CD40
|
CD40 molecule, TNF receptor superfamily member 5
|
|
chr7_+_128795178
|
3.115
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_149586341
|
3.110
|
NM_000449
NM_001025603
|
RFX5
|
regulatory factor X, 5 (influences HLA class II expression)
|
|
chr9_+_126460511
|
2.998
|
|
|
|
|
chr6_+_31647854
|
2.992
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr6_+_70633168
|
2.984
|
NM_001858
|
COL19A1
|
collagen, type XIX, alpha 1
|
|
chr11_+_303852
|
2.958
|
NM_003641
|
IFITM1
|
interferon induced transmembrane protein 1 (9-27)
|
|
chr7_+_73145481
|
2.864
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr7_+_128795280
|
2.845
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr19_+_17723285
|
2.835
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr5_-_169657312
|
2.809
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr19_+_17377502
|
2.734
|
|
LOC100507535
|
hypothetical LOC100507535
|
|
chr18_+_13601664
|
2.723
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr19_-_40925151
|
2.698
|
NM_024660
|
TMEM149
|
transmembrane protein 149
|
|
chr17_-_31231421
|
2.696
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr12_-_119961143
|
2.695
|
NM_003733
NM_198213
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr17_+_44643067
|
2.681
|
|
ABI3
|
ABI family, member 3
|
|
chr19_-_2036227
|
2.635
|
|
MOBKL2A
|
MOB1, Mps One Binder kinase activator-like 2A (yeast)
|
|
chr12_+_25096472
|
2.600
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_-_9777126
|
2.593
|
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr4_+_39874921
|
2.543
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr2_-_213723298
|
2.518
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr10_+_91082218
|
2.507
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr15_+_86983038
|
2.478
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr17_-_44642934
|
2.448
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr5_-_169657192
|
2.440
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_26665213
|
2.431
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_-_181889070
|
2.423
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr5_-_169657395
|
2.413
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr11_+_71387572
|
2.411
|
NM_001039659
NM_001039660
NM_173042
|
IL18BP
|
interleukin 18 binding protein
|
|
chr17_-_37518213
|
2.382
|
NM_024119
|
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58
|
|
chr19_-_44518465
|
2.377
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr22_+_38627031
|
2.367
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr19_-_44518516
|
2.317
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr2_-_215711313
|
2.313
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr1_-_159306327
|
2.281
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr3_+_190831909
|
2.235
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr5_-_169657360
|
2.216
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr6_+_29799182
|
2.213
|
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr5_-_149772563
|
2.194
|
|
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain
|
|
chr22_-_25205376
|
2.119
|
NM_152841
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr1_-_149004916
|
2.092
|
NM_004079
|
CTSS
|
cathepsin S
|
|
chr16_+_11966464
|
2.092
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr12_-_47604930
|
2.088
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr1_+_100957883
|
2.058
|
NM_001078
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr7_+_28418668
|
2.053
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr19_-_55104875
|
2.017
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr17_-_31441390
|
1.994
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr11_-_104411065
|
1.993
|
NM_001223
NM_033292
NM_033293
NM_033294
NM_033295
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr1_+_78888064
|
1.984
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr1_+_196874839
|
1.972
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_159306369
|
1.967
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr13_-_102517196
|
1.937
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr7_-_76667045
|
1.902
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr4_-_38482805
|
1.900
|
NM_003263
|
TLR1
|
toll-like receptor 1
|
|
chr1_-_153098811
|
1.882
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr3_-_49826221
|
1.881
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr16_+_55580910
|
1.872
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr17_+_6599879
|
1.868
|
NM_017523
NM_199139
|
XAF1
|
XIAP associated factor 1
|
|
chr11_+_5667494
|
1.850
|
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr1_+_40635058
|
1.834
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr16_-_88358946
|
1.821
|
|
FANCA
|
Fanconi anemia, complementation group A
|
|
chr12_+_11694172
|
1.775
|
|
ETV6
|
ets variant 6
|
|
chr1_+_159943385
|
1.757
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr13_-_45654327
|
1.736
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr22_+_38627129
|
1.720
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr11_+_71388310
|
1.703
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr3_-_58175437
|
1.678
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr4_+_113958652
|
1.610
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr12_-_47605478
|
1.603
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr4_+_113437947
|
1.597
|
NM_001102406
NM_025144
|
ALPK1
|
alpha-kinase 1
|
|
chr1_-_9052253
|
1.575
|
NM_001135585
NM_003039
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr10_+_35466715
|
1.554
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr6_-_138231006
|
1.547
|
|
|
|
|
chr2_-_37752731
|
1.522
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr17_+_55854636
|
1.514
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr6_-_149847718
|
1.510
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr17_+_7402788
|
1.501
|
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr4_-_153820535
|
1.493
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr12_+_6431437
|
1.487
|
NM_018009
|
TAPBPL
|
TAP binding protein-like
|
|
chr12_+_14409875
|
1.476
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr12_+_91620749
|
1.473
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr1_-_149004879
|
1.470
|
|
CTSS
|
cathepsin S
|
|
chr14_+_23700261
|
1.470
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr13_+_23742846
|
1.469
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr1_-_9052170
|
1.455
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr1_+_239762179
|
1.447
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chrX_+_37524190
|
1.440
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr19_+_40895669
|
1.432
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr19_-_17377383
|
1.425
|
NM_004335
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr6_-_76259976
|
1.395
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_196874759
|
1.395
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr3_-_114176401
|
1.383
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr8_-_145132582
|
1.381
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr7_+_150013716
|
1.381
|
NM_015660
|
GIMAP2
|
GTPase, IMAP family member 2
|
|
chr8_-_145132551
|
1.376
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr3_+_186563529
|
1.351
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chr17_-_26665144
|
1.349
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_+_938709
|
1.349
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr9_+_127550298
|
1.343
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr6_-_149847809
|
1.342
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr19_+_16691786
|
1.321
|
NM_001007525
|
NWD1
|
NACHT and WD repeat domain containing 1
|
|
chr4_-_169476391
|
1.314
|
NM_017631
|
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60
|
|
chr5_-_88155360
|
1.301
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr7_-_33047042
|
1.297
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr1_-_149004852
|
1.297
|
|
CTSS
|
cathepsin S
|
|
chr16_-_28425632
|
1.284
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr5_-_94442941
|
1.279
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr1_-_246869181
|
1.274
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chrX_+_37524250
|
1.271
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr10_-_7491204
|
1.253
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr7_+_119700924
|
1.231
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr3_-_3127057
|
1.231
|
NM_000564
NM_175724
NM_175725
NM_175726
NM_175727
NM_175728
|
IL5RA
|
interleukin 5 receptor, alpha
|
|
chr15_-_42790805
|
1.210
|
|
|
|
|
chr2_+_61986287
|
1.207
|
NM_152516
|
COMMD1
|
copper metabolism (Murr1) domain containing 1
|
|
chr10_-_1769669
|
1.202
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr1_+_154386358
|
1.201
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr1_+_32438788
|
1.194
|
NM_024296
|
CCDC28B
|
coiled-coil domain containing 28B
|
|
chr17_-_26665194
|
1.188
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr16_+_27346079
|
1.185
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr1_+_32512298
|
1.152
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr16_-_49272741
|
1.139
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr1_+_157441824
|
1.131
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr11_+_63060848
|
1.129
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr4_-_40211324
|
1.126
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr20_-_51633042
|
1.120
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr4_+_38740246
|
1.119
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr11_-_63808740
|
1.107
|
NM_004322
NM_032989
|
BAD
|
BCL2-associated agonist of cell death
|
|
chr22_-_16080265
|
1.106
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr2_+_182030510
|
1.106
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr17_-_39094787
|
1.099
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr1_+_84539876
|
1.086
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr11_+_71388619
|
1.080
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr8_+_26206648
|
1.080
|
NM_001177591
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr3_-_42789738
|
1.078
|
NM_144719
|
CCDC13
|
coiled-coil domain containing 13
|
|
chr17_-_26665220
|
1.072
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_+_5667392
|
1.068
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr20_-_5539487
|
1.059
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr6_-_30289092
|
1.057
|
NM_003449
|
TRIM26
|
tripartite motif containing 26
|
|
chr17_+_44642587
|
1.056
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr6_+_29799095
|
1.047
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr22_-_37480898
|
1.042
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr12_-_623056
|
1.037
|
|
|
|
|
chr9_+_5440455
|
1.037
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr12_+_46799279
|
1.031
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr3_-_15814678
|
1.028
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_47502115
|
1.026
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr13_+_42034871
|
1.026
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr4_-_87500201
|
1.022
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_-_25164061
|
1.014
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr3_-_145049853
|
1.013
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr2_-_191587127
|
1.008
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chr2_-_162883062
|
1.005
|
NM_022168
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr19_+_10057972
|
1.000
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr4_+_147316284
|
0.999
|
NM_007080
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr6_-_74284522
|
0.979
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_+_67404756
|
0.972
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr11_-_64327197
|
0.964
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr14_-_22358752
|
0.948
|
NM_001126106
|
SLC7A7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7
|
|
chr12_-_51887318
|
0.939
|
|
ITGB7
|
integrin, beta 7
|
|
chr12_-_119038995
|
0.921
|
NM_001167606
NM_006861
|
RAB35
|
RAB35, member RAS oncogene family
|
|
chr13_+_107719977
|
0.911
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr4_-_116254381
|
0.904
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr15_-_27349233
|
0.890
|
|
NDNL2
|
necdin-like 2
|