|
chr7_+_50318801
|
5.959
|
|
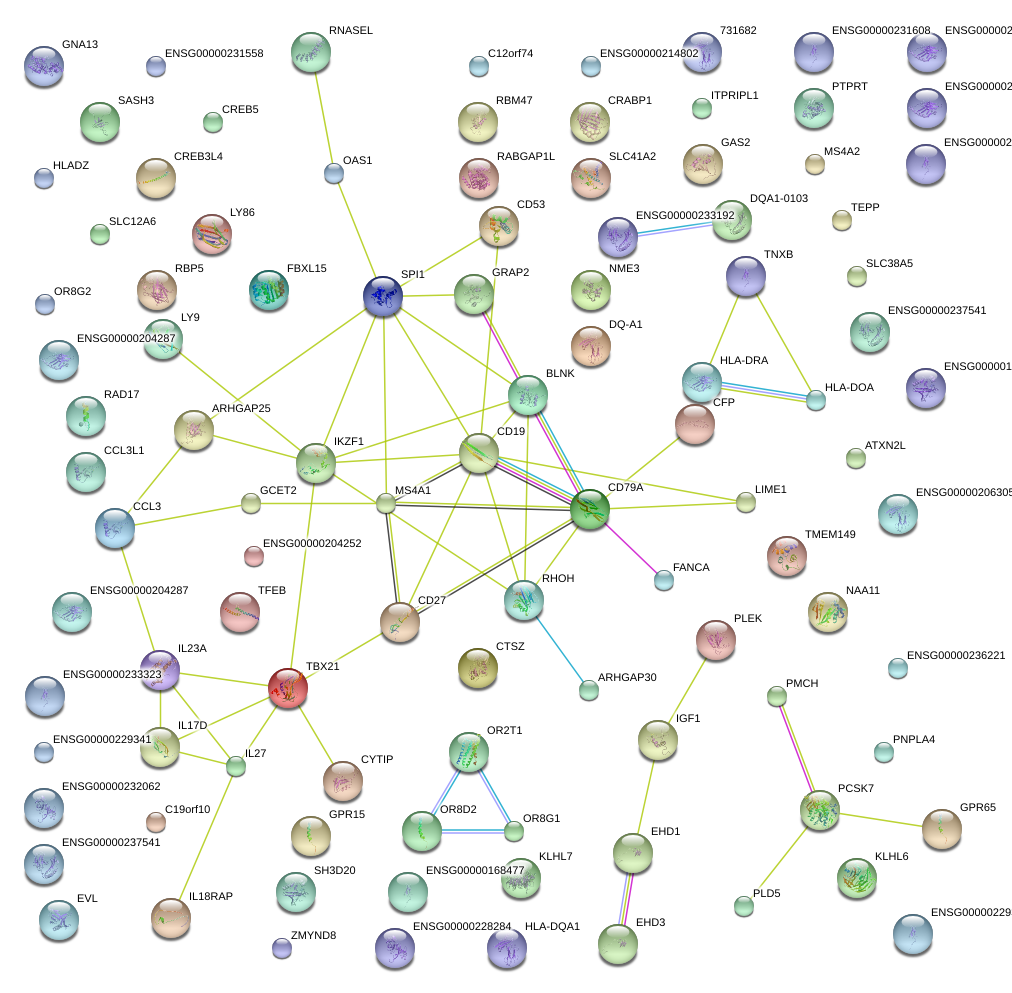
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr3_+_188197029
|
4.606
|
|
|
|
|
chr10_-_98021144
|
4.355
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_102401580
|
3.937
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr12_-_7172641
|
3.412
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr2_-_158008763
|
3.353
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr1_+_173111306
|
3.313
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr4_-_40211324
|
3.099
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr20_-_57015698
|
3.017
|
NM_001336
|
CTSZ
|
cathepsin Z
|
|
chr4_+_39874921
|
2.924
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr15_-_32416336
|
2.886
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr11_+_59979857
|
2.840
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr22_+_38627031
|
2.762
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr11_+_123600553
|
2.724
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr2_+_96355661
|
2.713
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr17_-_31441390
|
2.632
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chrX_+_65152004
|
2.520
|
|
|
|
|
chr20_-_45410033
|
2.459
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr20_-_57015547
|
2.446
|
|
CTSZ
|
cathepsin Z
|
|
chr12_+_6424293
|
2.425
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chrX_-_48212683
|
2.386
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr4_-_80466177
|
2.351
|
NM_032693
|
NAA11
|
N(alpha)-acetyltransferase 11, NatA catalytic subunit
|
|
chr17_-_40843530
|
2.291
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr6_+_32515596
|
2.291
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr14_+_87541220
|
2.123
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr3_+_99733432
|
2.107
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr12_+_91620749
|
2.073
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr6_-_33085290
|
2.062
|
|
HLA-DOA
|
major histocompatibility complex, class II, DO alpha
|
|
chr11_-_47356650
|
2.061
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr22_+_38627129
|
2.031
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr11_-_47356562
|
1.993
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47356642
|
1.927
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr1_+_111217244
|
1.886
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr11_-_47356672
|
1.871
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr16_+_56567839
|
1.838
|
NM_199046
NM_199456
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr12_-_101396459
|
1.747
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_-_103846186
|
1.727
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr6_-_33085329
|
1.711
|
NM_002119
|
HLA-DOA
|
major histocompatibility complex, class II, DO alpha
|
|
chr12_+_111829191
|
1.688
|
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr16_-_88358946
|
1.687
|
|
FANCA
|
Fanconi anemia, complementation group A
|
|
chr19_+_47073029
|
1.671
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chrX_-_7855400
|
1.616
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr12_+_55018925
|
1.609
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr11_+_22652957
|
1.600
|
|
GAS2
|
growth arrest-specific 2
|
|
chr1_+_159032551
|
1.593
|
NM_001033667
NM_002348
|
LY9
|
lymphocyte antigen 9
|
|
chrX_-_47374647
|
1.586
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr17_-_53764867
|
1.567
|
|
|
|
|
chr6_-_41811916
|
1.552
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr17_-_60439764
|
1.518
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr16_-_1761504
|
1.514
|
|
NME3
|
non-metastatic cells 3, protein expressed in
|
|
chr6_-_32121784
|
1.506
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr1_-_159306037
|
1.485
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr17_+_43165608
|
1.485
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr11_-_47356479
|
1.436
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr3_-_113334657
|
1.435
|
NM_001190259
NM_001190260
NM_152785
|
GCET2
|
germinal center expressed transcript 2
|
|
chr20_+_61838479
|
1.428
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr2_+_68445936
|
1.416
|
|
PLEK
|
pleckstrin
|
|
chr11_+_22652894
|
1.396
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr6_+_6533932
|
1.394
|
NM_004271
|
LY86
|
lymphocyte antigen 86
|
|
chr1_-_180822686
|
1.387
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr12_-_101115677
|
1.377
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr7_+_28418668
|
1.374
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_+_123639909
|
1.366
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr19_-_40925151
|
1.339
|
NM_024660
|
TMEM149
|
transmembrane protein 149
|
|
chr1_+_246635918
|
1.337
|
NM_030904
|
OR2T1
|
olfactory receptor, family 2, subfamily T, member 1
|
|
chr11_-_64403712
|
1.336
|
|
EHD1
|
EH-domain containing 1
|
|
chr11_+_123625632
|
1.327
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr11_-_116605761
|
1.326
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chrX_+_128741578
|
1.323
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr1_-_240679380
|
1.302
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr1_+_152206969
|
1.287
|
NM_130898
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4
|
|
chr14_+_99601498
|
1.271
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr16_-_28425632
|
1.267
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr3_-_184756101
|
1.247
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr2_+_68855436
|
1.226
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr16_+_28850760
|
1.203
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr5_+_68703036
|
1.200
|
NM_133339
|
RAD17
|
RAD17 homolog (S. pombe)
|
|
chr20_+_61838389
|
1.198
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr10_+_104169560
|
1.196
|
NM_024326
|
FBXL15
|
F-box and leucine-rich repeat protein 15
|
|
chr6_+_32713160
|
1.191
|
NM_002122
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1
|
|
chr1_-_218329684
|
1.183
|
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chr1_+_154390939
|
1.176
|
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chrX_-_130792297
|
1.170
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr2_-_65013070
|
1.164
|
|
LOC400958
|
hypothetical LOC400958
|
|
chr6_-_25834733
|
1.142
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr3_-_122862391
|
1.140
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr10_-_104169458
|
1.120
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr7_-_38355949
|
1.119
|
|
|
|
|
chr1_-_218329773
|
1.108
|
NM_006085
|
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1
|
|
chr1_+_144124547
|
1.106
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr19_-_44518465
|
1.103
|
|
GMFG
|
glia maturation factor, gamma
|
|
chrX_+_128741562
|
1.101
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_-_20340377
|
1.100
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr18_-_28247193
|
1.091
|
|
|
|
|
chr8_-_107023
|
1.066
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr7_+_106897737
|
1.065
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_+_111544469
|
1.061
|
|
|
|
|
chr1_-_114215768
|
1.054
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr16_-_49272617
|
1.054
|
|
SNX20
|
sorting nexin 20
|
|
chr1_+_43063817
|
1.048
|
NM_018538
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group)
|
|
chr9_-_97308687
|
1.042
|
|
PTCH1
|
patched 1
|
|
chrX_+_65300803
|
1.032
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr17_-_36594652
|
1.032
|
NM_033060
|
KRTAP4-1
|
keratin associated protein 4-1
|
|
chr5_+_132021763
|
1.027
|
NM_002188
|
IL13
|
interleukin 13
|
|
chr3_-_121884091
|
1.026
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr1_-_152185764
|
1.022
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr17_-_53946899
|
1.021
|
|
MTMR4
|
myotubularin related protein 4
|
|
chr16_-_49272741
|
1.018
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr11_+_5367182
|
1.013
|
NM_001004756
|
OR51M1
|
olfactory receptor, family 51, subfamily M, member 1
|
|
chr18_-_28247186
|
0.995
|
|
|
|
|
chr1_-_148854503
|
0.989
|
|
|
|
|
chr12_-_51585004
|
0.988
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr6_-_30188845
|
0.987
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr1_+_157526127
|
0.972
|
NM_002001
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide
|
|
chr15_+_42815851
|
0.964
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr14_+_59782222
|
0.960
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr14_+_77939845
|
0.959
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr16_+_49288550
|
0.952
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr1_+_210275735
|
0.949
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr13_+_110637173
|
0.948
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr19_-_44518516
|
0.945
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr10_+_70152216
|
0.944
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr6_-_76022565
|
0.943
|
|
TMEM30A
|
transmembrane protein 30A
|
|
chr1_+_210275752
|
0.941
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr17_-_36209672
|
0.939
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr3_-_122862447
|
0.937
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr19_-_19487237
|
0.920
|
NM_032037
|
TSSK6
|
testis-specific serine kinase 6
|
|
chr14_+_99601517
|
0.898
|
|
EVL
|
Enah/Vasp-like
|
|
chr11_-_123918759
|
0.889
|
NM_001005195
|
OR8B12
|
olfactory receptor, family 8, subfamily B, member 12
|
|
chr1_+_25471470
|
0.883
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr6_+_14032655
|
0.880
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr1_-_24067347
|
0.880
|
NM_000147
|
FUCA1
|
fucosidase, alpha-L- 1, tissue
|
|
chr2_+_191009265
|
0.876
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chrX_+_134803450
|
0.872
|
NM_018666
|
SAGE1
|
sarcoma antigen 1
|
|
chr15_+_89248423
|
0.872
|
NM_006122
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chrX_+_9391314
|
0.867
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr6_-_137536412
|
0.864
|
NM_052962
NM_181309
NM_181310
|
IL22RA2
|
interleukin 22 receptor, alpha 2
|
|
chr15_-_78050515
|
0.861
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_-_153098811
|
0.860
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_+_557011
|
0.836
|
NM_176677
|
NHLRC4
PIGQ
|
NHL repeat containing 4
phosphatidylinositol glycan anchor biosynthesis, class Q
|
|
chr14_-_20340830
|
0.835
|
NM_002933
NM_198234
NM_198235
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr1_-_159306369
|
0.827
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr9_-_70780574
|
0.825
|
|
|
|
|
chr16_+_29581783
|
0.820
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr1_-_159306327
|
0.809
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr8_-_66863841
|
0.807
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr12_-_9777126
|
0.804
|
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr1_-_210275402
|
0.802
|
NM_015434
|
INTS7
|
integrator complex subunit 7
|
|
chr7_-_25234582
|
0.797
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr8_+_144802972
|
0.795
|
NM_014789
|
ZNF623
|
zinc finger protein 623
|
|
chr6_+_106653429
|
0.789
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr14_-_52689782
|
0.787
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr19_-_59309719
|
0.780
|
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr2_+_183290912
|
0.780
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr6_+_41411505
|
0.778
|
NM_004828
|
NCR2
|
natural cytotoxicity triggering receptor 2
|
|
chr12_-_56159895
|
0.776
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr5_+_180190585
|
0.767
|
|
LOC729678
|
hypothetical LOC729678
|
|
chr14_-_22658078
|
0.764
|
NM_001805
|
CEBPE
|
CCAAT/enhancer binding protein (C/EBP), epsilon
|
|
chr21_+_44597911
|
0.763
|
NM_003307
|
TRPM2
|
transient receptor potential cation channel, subfamily M, member 2
|
|
chr2_-_171725150
|
0.759
|
|
TLK1
|
tousled-like kinase 1
|
|
chr1_-_226357552
|
0.759
|
NM_024319
|
C1orf35
|
chromosome 1 open reading frame 35
|
|
chr10_+_5976354
|
0.757
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr1_+_210275541
|
0.757
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr12_+_111829121
|
0.754
|
NM_001032409
NM_002534
NM_016816
|
OAS1
|
2',5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr17_+_44643067
|
0.754
|
|
ABI3
|
ABI family, member 3
|
|
chr9_+_95886558
|
0.753
|
NM_152422
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1
|
|
chr12_+_6925997
|
0.750
|
NM_080548
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr4_-_38710376
|
0.749
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr11_-_55661769
|
0.747
|
NM_001004064
|
OR8J3
|
olfactory receptor, family 8, subfamily J, member 3
|
|
chr13_-_45654296
|
0.739
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_-_123999640
|
0.732
|
NM_006073
|
TRDN
|
triadin
|
|
chr16_-_79811475
|
0.721
|
NM_001076780
NM_052892
|
PKD1L2
|
polycystic kidney disease 1-like 2
|
|
chr1_-_47468029
|
0.720
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr4_+_71235259
|
0.716
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr3_-_25658829
|
0.714
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr13_+_52114565
|
0.707
|
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2
|
|
chr1_+_160618143
|
0.705
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr6_-_159386007
|
0.704
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr4_-_116254381
|
0.701
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr6_+_14225836
|
0.696
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr13_-_45654292
|
0.693
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_+_35373632
|
0.685
|
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr15_+_89246780
|
0.684
|
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2
|
|
chr2_-_142605010
|
0.678
|
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chrX_-_70390666
|
0.675
|
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr22_+_33792125
|
0.672
|
NM_001008494
|
ISX
|
intestine-specific homeobox
|
|
chr12_+_6926014
|
0.670
|
|
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6
|
|
chr3_+_52788976
|
0.669
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr7_+_106293112
|
0.669
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr14_-_23107106
|
0.664
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr13_-_45654327
|
0.652
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr3_+_120375053
|
0.641
|
NM_006952
|
UPK1B
|
uroplakin 1B
|
|
chr11_+_303852
|
0.640
|
NM_003641
|
IFITM1
|
interferon induced transmembrane protein 1 (9-27)
|
|
chr20_-_34807882
|
0.638
|
NM_022477
NM_032013
|
NDRG3
|
NDRG family member 3
|
|
chr17_-_44642934
|
0.638
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr6_-_30760413
|
0.638
|
|
KIAA1949
|
KIAA1949
|
|
chr5_-_135729062
|
0.637
|
NM_001167576
NM_001167577
NM_020389
|
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7
|
|
chr7_-_100077072
|
0.636
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chrX_-_70390704
|
0.632
|
NM_201599
|
ZMYM3
|
zinc finger, MYM-type 3
|
|
chr11_-_123685871
|
0.631
|
NM_001002917
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1
|