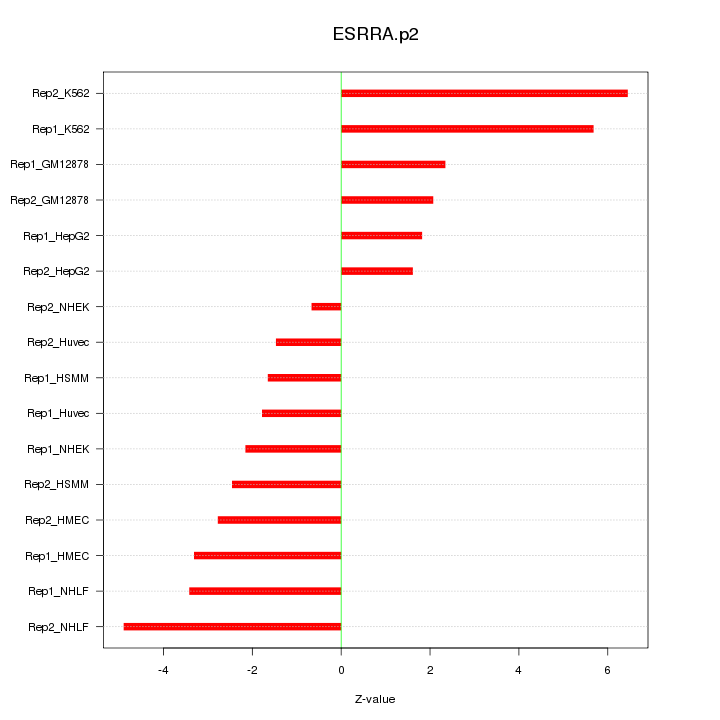
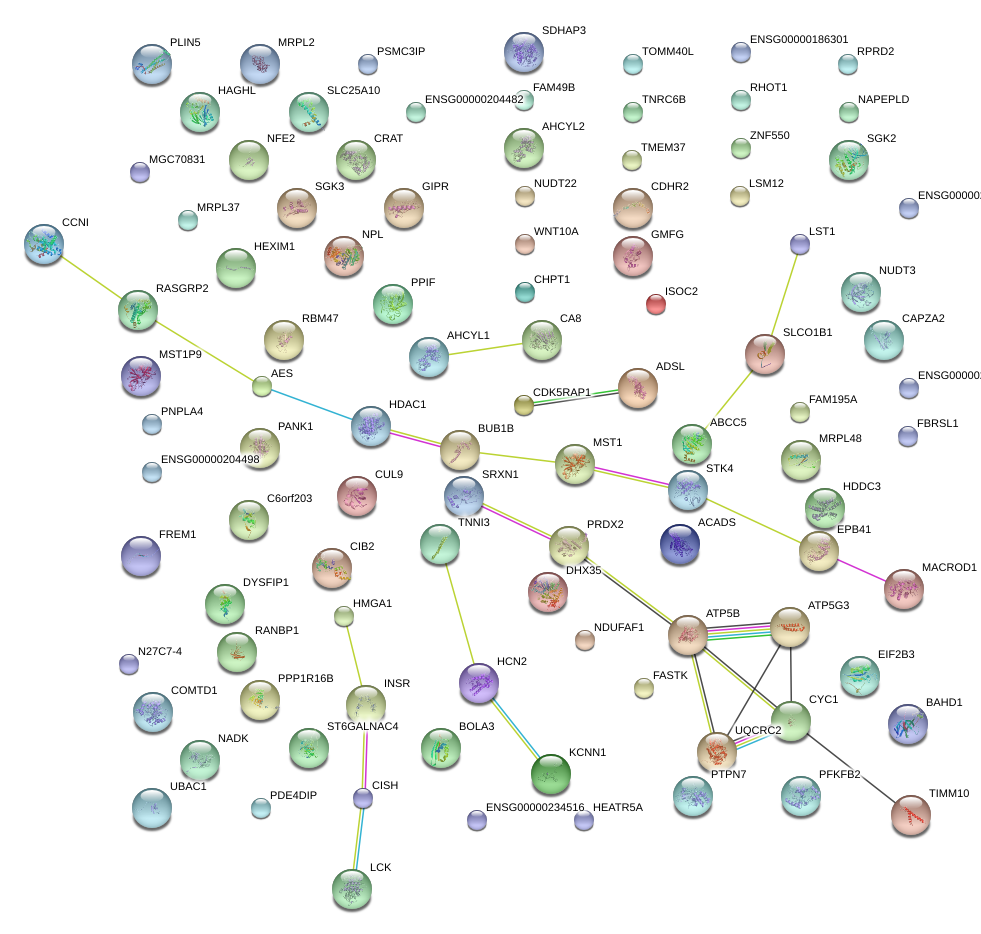
|
chr1_-_143706265
|
8.301
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr1_-_143706196
|
6.195
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr22_-_22440054
|
5.295
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr2_-_175754366
|
5.120
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr1_+_29086206
|
4.831
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr2_+_119905812
|
4.530
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr1_+_29086156
|
4.529
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr12_-_52980984
|
4.366
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr12_-_52980914
|
4.194
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr6_+_34314545
|
4.123
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr3_-_49701085
|
3.939
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr19_-_44518465
|
3.750
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr15_-_89276657
|
3.523
|
NM_198527
|
HDDC3
|
HD domain containing 3
|
|
chr4_-_40212670
|
3.372
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr19_-_44518516
|
3.223
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr12_-_55325912
|
3.221
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr5_-_1647617
|
3.063
|
|
SDHAP3
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3
|
|
chr20_+_37024383
|
3.059
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr19_-_4486202
|
3.032
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chrX_+_65152004
|
3.029
|
|
|
|
|
chr20_+_36867708
|
2.970
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr16_+_631902
|
2.771
|
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr11_+_73176561
|
2.722
|
NM_016055
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr19_-_12773591
|
2.721
|
NM_005809
NM_181738
|
PRDX2
|
peroxiredoxin 2
|
|
chr10_-_91394881
|
2.693
|
|
PANK1
|
pantothenate kinase 1
|
|
chr6_+_44076281
|
2.595
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr11_-_57054756
|
2.589
|
NM_012456
|
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast)
|
|
chr15_+_38520594
|
2.554
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr12_+_100615530
|
2.524
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr22_+_22703107
|
2.512
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chr16_+_631828
|
2.496
|
NM_138418
|
FAM195A
|
family with sequence similarity 195, member A
|
|
chr2_-_74228506
|
2.458
|
NM_001035505
NM_212552
|
BOLA3
|
bolA homolog 3 (E. coli)
|
|
chr10_-_76665691
|
2.437
|
NM_144589
|
COMTD1
|
catechol-O-methyltransferase domain containing 1
|
|
chr22_+_18485145
|
2.412
|
|
RANBP1
|
RAN binding protein 1
|
|
chr22_+_18485095
|
2.411
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_-_1679940
|
2.407
|
NM_001198995
|
NADK
|
NAD kinase
|
|
chr1_+_148603769
|
2.396
|
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr1_-_45224784
|
2.390
|
|
EIF2B3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma, 58kDa
|
|
chr16_+_21872109
|
2.335
|
NM_003366
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr20_+_41621119
|
2.325
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr5_+_175902117
|
2.320
|
NM_001171976
|
CDHR2
|
cadherin-related family member 2
|
|
chr11_-_64268841
|
2.288
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr1_+_32530298
|
2.285
|
|
HDAC1
|
histone deacetylase 1
|
|
chr22_+_18485076
|
2.285
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_+_32530289
|
2.277
|
|
HDAC1
|
histone deacetylase 1
|
|
chr1_+_32530294
|
2.265
|
NM_004964
|
HDAC1
|
histone deacetylase 1
|
|
chr19_-_3013441
|
2.260
|
|
AES
|
amino-terminal enhancer of split
|
|
chr22_+_38903825
|
2.226
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_-_50624085
|
2.224
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr20_+_41621079
|
2.211
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr2_+_219453487
|
2.194
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr16_+_21872201
|
2.178
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr16_+_21872178
|
2.160
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr12_-_55326043
|
2.152
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr16_+_21872155
|
2.147
|
|
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II
|
|
chr22_+_18485068
|
2.112
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_-_3013191
|
2.104
|
NM_001130
NM_198970
|
AES
|
amino-terminal enhancer of split
|
|
chr10_+_80777265
|
2.092
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr15_+_38240555
|
2.075
|
|
BUB1B
|
budding uninhibited by benzimidazoles 1 homolog beta (yeast)
|
|
chr1_-_143706300
|
2.074
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr7_-_102576570
|
2.065
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr15_-_39481903
|
2.054
|
NM_016013
|
NDUFAF1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1
|
|
chr10_+_80777248
|
2.028
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr6_-_43135092
|
2.004
|
NM_015950
|
MRPL2
|
mitochondrial ribosomal protein L2
|
|
chr12_-_55326052
|
2.001
|
NM_001686
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr11_-_63689780
|
1.984
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr12_+_119647953
|
1.953
|
NM_000017
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain
|
|
chr3_-_185218311
|
1.925
|
NM_001023587
NM_005688
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5
|
|
chr19_+_540849
|
1.920
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr17_+_40580466
|
1.871
|
NM_006460
|
HEXIM1
|
hexamethylene bis-acetamide inducible 1
|
|
chr11_+_63750296
|
1.841
|
NM_001128612
NM_001128613
NM_032344
|
NUDT22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22
|
|
chr17_+_27493680
|
1.837
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr1_+_205293217
|
1.826
|
NM_001018053
NM_006212
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr7_+_128795280
|
1.822
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr20_-_581808
|
1.814
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr16_-_30694799
|
1.811
|
|
|
|
|
chr6_-_34468076
|
1.806
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr7_+_116238309
|
1.802
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr9_-_130912728
|
1.795
|
NM_000755
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr20_-_581844
|
1.789
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr19_-_62759536
|
1.787
|
NM_001039654
|
ZNF550
|
zinc finger protein 550
|
|
chr14_+_87560646
|
1.778
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr15_-_76210611
|
1.763
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr16_+_716936
|
1.762
|
NM_207112
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr1_+_32489426
|
1.757
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr14_-_30959487
|
1.754
|
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chrX_-_7855764
|
1.733
|
NM_001142389
NM_001172672
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr19_+_17923110
|
1.731
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr19_-_60664860
|
1.727
|
NM_001136201
NM_001136202
NM_024710
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr22_+_18484810
|
1.727
|
NM_002882
|
RANBP1
|
RAN binding protein 1
|
|
chr22_+_18485057
|
1.706
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_-_60360768
|
1.703
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr1_-_203985932
|
1.698
|
|
|
|
|
chr17_-_77386156
|
1.695
|
|
DYSFIP1
|
dysferlin interacting protein 1
|
|
chr6_+_31662454
|
1.671
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr17_-_77386212
|
1.667
|
NM_001007533
|
DYSFIP1
|
dysferlin interacting protein 1
|
|
chr6_+_107456020
|
1.659
|
NM_001142468
NM_001142470
NM_016487
|
C6orf203
|
chromosome 6 open reading frame 203
|
|
chr19_+_50863341
|
1.644
|
NM_000164
|
GIPR
|
gastric inhibitory polypeptide receptor
|
|
chr8_-_61356436
|
1.638
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr20_-_31452964
|
1.626
|
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr9_-_14858974
|
1.621
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr7_-_38355949
|
1.615
|
|
|
|
|
chr1_-_200396354
|
1.614
|
NM_002832
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr17_+_27493618
|
1.601
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr17_-_39500512
|
1.600
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr17_+_27493654
|
1.581
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr8_-_131097856
|
1.572
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_+_181028016
|
1.571
|
NM_030769
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr8_+_145221973
|
1.560
|
|
CYC1
|
cytochrome c-1
|
|
chr19_-_7244876
|
1.560
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr7_-_150408824
|
1.558
|
NM_006712
NM_033015
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr19_-_60664809
|
1.557
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr8_+_145221924
|
1.541
|
NM_001916
|
CYC1
|
cytochrome c-1
|
|
chr6_+_43257883
|
1.535
|
NM_015089
|
CUL9
|
cullin 9
|
|
chr12_+_131577229
|
1.532
|
NM_001142641
|
FBRSL1
|
fibrosin-like 1
|
|
chr1_+_159462456
|
1.526
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr17_-_37983175
|
1.522
|
NM_013290
NM_016556
|
PSMC3IP
|
PSMC3 interacting protein
|
|
chr9_-_129719031
|
1.521
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr17_+_77289775
|
1.512
|
NM_012140
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr9_-_137992766
|
1.508
|
|
UBAC1
|
UBA domain containing 1
|
|
chr1_-_203985963
|
1.508
|
NM_022731
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr15_-_64577122
|
1.498
|
NM_006049
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa
|
|
chr9_-_137992967
|
1.494
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr19_+_10515591
|
1.489
|
NM_032885
|
ATG4D
|
ATG4 autophagy related 4 homolog D (S. cerevisiae)
|
|
chr11_+_62380042
|
1.484
|
NM_001012662
NM_001012664
NM_002394
NM_001012661
NM_001012663
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr8_-_131097828
|
1.477
|
|
FAM49B
|
family with sequence similarity 49, member B
|
|
chr1_+_234625338
|
1.474
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr7_+_2409731
|
1.466
|
NM_018641
|
CHST12
|
carbohydrate (chondroitin 4) sulfotransferase 12
|
|
chr11_-_63749916
|
1.460
|
NM_001160393
|
TRPT1
|
tRNA phosphotransferase 1
|
|
chr20_-_31452991
|
1.426
|
NM_016082
NM_016408
|
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1
|
|
chr6_-_97452416
|
1.425
|
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
|
|
chr19_-_12905459
|
1.422
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr7_+_128795178
|
1.419
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr7_-_38356100
|
1.412
|
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr10_-_27026559
|
1.401
|
|
|
|
|
chr17_+_77289804
|
1.386
|
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10
|
|
chr19_-_12905524
|
1.378
|
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr7_-_150408785
|
1.374
|
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr8_+_145221966
|
1.374
|
|
CYC1
|
cytochrome c-1
|
|
chr19_-_60664747
|
1.374
|
|
ISOC2
|
isochorismatase domain containing 2
|
|
chr4_-_38460983
|
1.370
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr3_-_122862391
|
1.359
|
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chrX_+_153310313
|
1.352
|
|
ATP6AP1
|
ATPase, H+ transporting, lysosomal accessory protein 1
|
|
chr2_+_176842368
|
1.350
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr22_+_35586950
|
1.350
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr19_-_45947667
|
1.336
|
NM_198476
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr16_+_31274020
|
1.333
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr11_-_3818713
|
1.325
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr20_-_44713466
|
1.316
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr11_-_63750138
|
1.301
|
NM_001033678
NM_001160389
NM_001160390
NM_001160392
NM_031472
|
TRPT1
|
tRNA phosphotransferase 1
|
|
chr12_-_116021473
|
1.289
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr11_+_22652957
|
1.285
|
|
GAS2
|
growth arrest-specific 2
|
|
chr1_+_55237193
|
1.285
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr2_+_228386800
|
1.280
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr9_-_99994704
|
1.278
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr19_-_12905528
|
1.277
|
NM_004461
|
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit
|
|
chr1_+_29113589
|
1.275
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr4_-_74307637
|
1.274
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr17_+_27493486
|
1.270
|
NM_001033566
NM_001033568
NM_018307
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr9_-_130912603
|
1.268
|
|
CRAT
|
carnitine O-acetyltransferase
|
|
chr7_-_140270749
|
1.266
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr11_+_47227009
|
1.257
|
NM_001130102
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr11_-_65895640
|
1.254
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr5_-_126437070
|
1.250
|
NM_001164478
NM_001164479
|
FLJ44606
|
glutaredoxin-like protein YDR286C homolog
|
|
chr1_-_27689129
|
1.247
|
|
WASF2
|
WAS protein family, member 2
|
|
chr18_+_42751510
|
1.244
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr9_-_139214962
|
1.236
|
NM_001128228
|
TPRN
|
taperin
|
|
chr19_-_3013768
|
1.235
|
|
AES
|
amino-terminal enhancer of split
|
|
chrX_-_153397566
|
1.235
|
NM_001171132
NM_001171133
NM_001171134
NM_021806
|
FAM3A
|
family with sequence similarity 3, member A
|
|
chr1_-_199259447
|
1.226
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr18_-_648246
|
1.207
|
NM_001012716
|
C18orf56
|
chromosome 18 open reading frame 56
|
|
chr7_-_122985060
|
1.204
|
NM_005000
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa
|
|
chr1_-_172153087
|
1.203
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr6_+_37081387
|
1.199
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr11_-_86343859
|
1.192
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr19_-_13074973
|
1.185
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr6_-_43592670
|
1.184
|
|
YIPF3
|
Yip1 domain family, member 3
|
|
chr4_+_2440977
|
1.183
|
NM_001185009
|
RNF4
|
ring finger protein 4
|
|
chr19_-_1430194
|
1.181
|
|
C19orf25
|
chromosome 19 open reading frame 25
|
|
chr8_-_100975086
|
1.180
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr1_-_200396328
|
1.178
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr6_-_34468288
|
1.175
|
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr19_-_1430116
|
1.168
|
|
C19orf25
|
chromosome 19 open reading frame 25
|
|
chr8_-_100975041
|
1.168
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr2_-_26320967
|
1.164
|
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit
|
|
chr6_+_107456457
|
1.164
|
|
C6orf203
|
chromosome 6 open reading frame 203
|
|
chr10_+_22645370
|
1.158
|
|
COMMD3
|
COMM domain containing 3
|
|
chr10_+_80777234
|
1.154
|
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr12_-_642786
|
1.153
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr2_+_112956192
|
1.152
|
|
TTL
|
tubulin tyrosine ligase
|
|
chr10_+_80777199
|
1.134
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr7_+_94374862
|
1.134
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr1_+_47572075
|
1.131
|
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic
|
|
chrX_+_65299157
|
1.127
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr16_+_76313909
|
1.126
|
NM_001105663
|
NUDT7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7
|
|
chr3_-_42820915
|
1.123
|
NM_001099668
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A
|
|
chr17_-_70430839
|
1.122
|
NM_173477
|
USH1G
|
Usher syndrome 1G (autosomal recessive)
|
|
chrX_+_48505097
|
1.119
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr14_-_90352395
|
1.116
|
|
|
|
|
chr10_+_22645304
|
1.113
|
NM_012071
|
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 read-through transcript
COMM domain containing 3
|