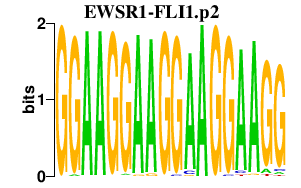
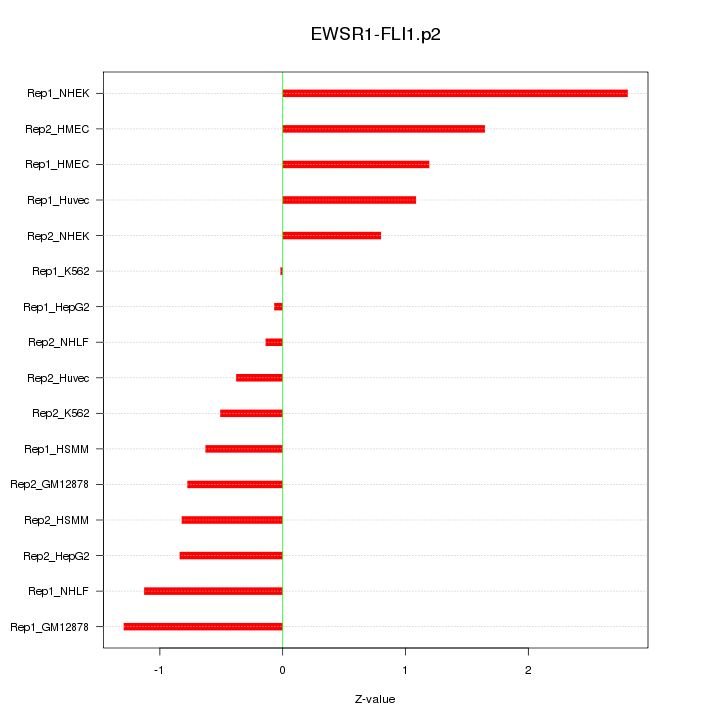
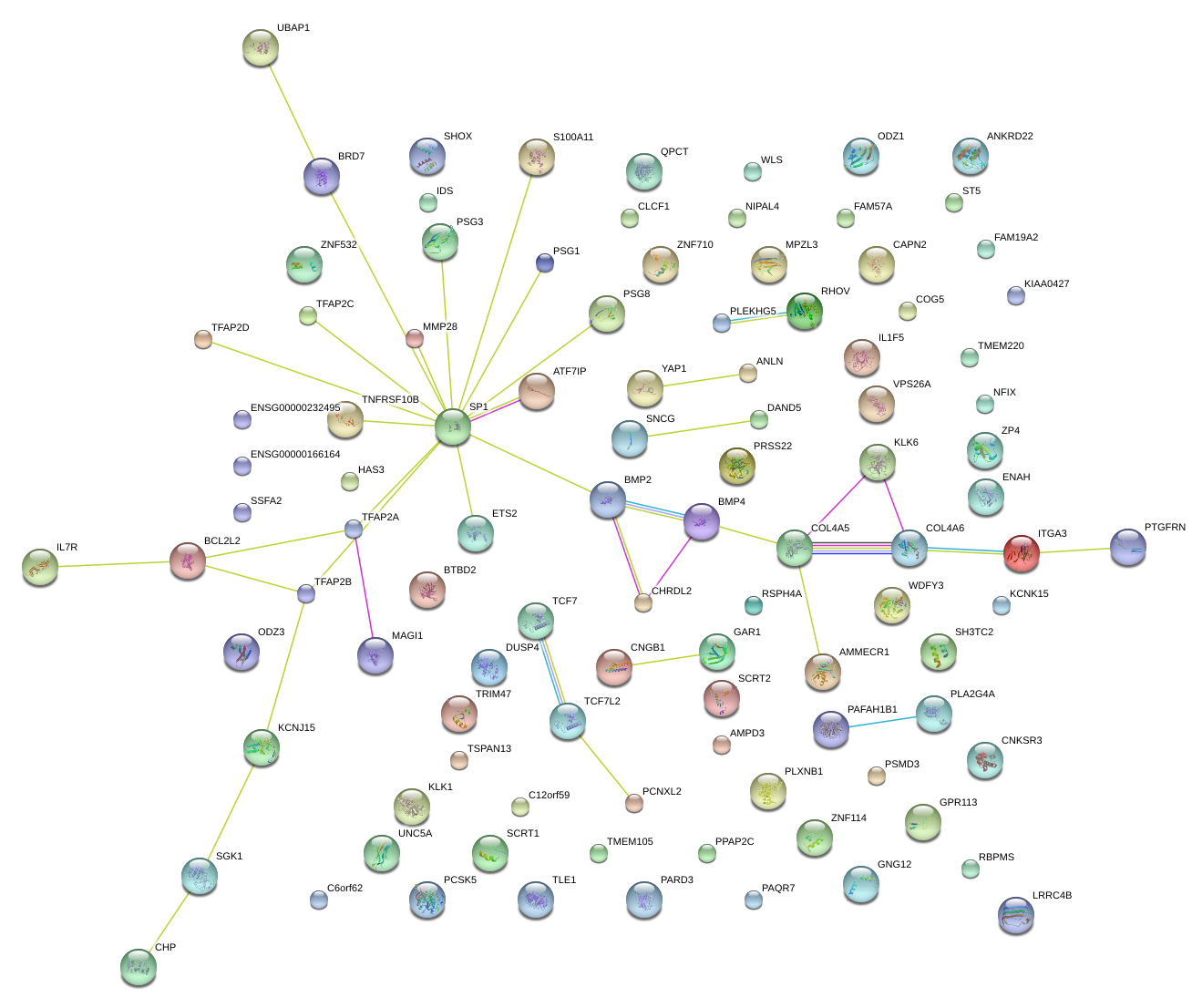
|
chr6_-_10520264
|
4.478
|
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr6_-_10520592
|
4.071
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr5_-_148422799
|
2.472
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr4_+_183302105
|
2.065
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr1_-_68071635
|
1.956
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr11_-_117628211
|
1.941
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chrX_+_107569671
|
1.900
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chrX_-_148394570
|
1.526
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr11_+_101485946
|
1.515
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr7_+_16759845
|
1.487
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr1_-_6468600
|
1.295
|
NM_001042665
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr16_+_67697622
|
1.290
|
NM_138612
|
HAS3
|
hyaluronan synthase 3
|
|
chr17_-_31146660
|
1.276
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chr1_-_150276064
|
1.273
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr1_-_150276091
|
1.186
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr1_-_150276114
|
1.184
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr1_-_68470585
|
1.169
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr7_-_106991941
|
1.145
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chrX_-_107569256
|
1.141
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr3_-_65999548
|
1.120
|
NM_001033057
NM_004742
NM_015520
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr9_+_77695334
|
1.099
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr17_+_2443983
|
1.068
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr18_+_44319365
|
1.059
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr10_+_114700775
|
1.052
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr17_+_45488356
|
1.007
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr17_+_45488330
|
0.990
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr1_-_223907283
|
0.934
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_+_113532685
|
0.930
|
NM_173170
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr17_+_2443652
|
0.922
|
NM_000430
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr19_-_242168
|
0.905
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr2_+_37425240
|
0.897
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr5_+_156819782
|
0.876
|
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr7_+_36395913
|
0.862
|
NM_018685
|
ANLN
|
anillin, actin binding protein
|
|
chr21_+_39099624
|
0.831
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr17_-_71385651
|
0.825
|
|
TRIM47
|
tripartite motif containing 47
|
|
chr10_+_88708267
|
0.819
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr1_+_221955876
|
0.817
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr20_+_6696744
|
0.817
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr1_+_185064654
|
0.812
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr1_-_236120844
|
0.808
|
NM_021186
|
ZP4
|
zona pellucida glycoprotein 4
|
|
chr4_-_86106567
|
0.798
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr19_+_12996830
|
0.798
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_-_231498081
|
0.797
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|
|
chr6_+_139055244
|
0.788
|
|
FLJ46906
|
hypothetical LOC441172
|
|
chr16_-_48960186
|
0.777
|
NM_001173984
NM_013263
|
BRD7
|
bromodomain containing 7
|
|
chr21_+_39099717
|
0.747
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr9_-_83494198
|
0.733
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr3_-_48446405
|
0.694
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr19_-_55763091
|
0.694
|
NM_001080457
|
LRRC4B
|
leucine rich repeat containing 4B
|
|
chr18_+_54682261
|
0.683
|
|
ZNF532
|
zinc finger protein 532
|
|
chr2_+_113533155
|
0.664
|
NM_012275
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr8_-_145530639
|
0.633
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr12_+_14409875
|
0.628
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr19_-_1966628
|
0.624
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr19_+_12996745
|
0.595
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_53466465
|
0.586
|
NM_153608
|
ZNF114
|
zinc finger protein 114
|
|
chr2_-_26395420
|
0.586
|
NM_001145168
NM_001145169
|
GPR113
|
G protein-coupled receptor 113
|
|
chr11_+_9736408
|
0.571
|
|
LOC283104
|
hypothetical LOC283104
|
|
chr8_+_30361485
|
0.562
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr14_-_53493198
|
0.558
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr16_-_2848106
|
0.557
|
NM_022119
|
PRSS22
|
protease, serine, 22
|
|
chr10_+_119292699
|
0.557
|
|
|
|
|
chr1_-_72521079
|
0.540
|
|
|
|
|
chr15_+_39310693
|
0.515
|
NM_007236
|
CHP
|
calcium binding protein P22
|
|
chr15_+_39310757
|
0.515
|
|
CHP
|
calcium binding protein P22
|
|
chr17_-_10574370
|
0.512
|
NM_001004313
|
TMEM220
|
transmembrane protein 220
|
|
chr6_-_24827265
|
0.495
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr17_+_35390607
|
0.495
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr19_-_242335
|
0.489
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr21_+_38550739
|
0.471
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr21_+_38550533
|
0.470
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr20_+_42807901
|
0.469
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr10_-_35143900
|
0.468
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr17_+_35390614
|
0.467
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr2_+_182464910
|
0.463
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr10_-_90601655
|
0.462
|
NM_144590
|
ANKRD22
|
ankyrin repeat domain 22
|
|
chr8_-_22982435
|
0.443
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr10_+_70553993
|
0.439
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_+_117253882
|
0.430
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr15_+_88345612
|
0.414
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr9_+_34169023
|
0.414
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr5_+_156819604
|
0.413
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr1_-_155336223
|
0.409
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr17_+_35390579
|
0.407
|
NM_002809
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr14_+_22845851
|
0.398
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr9_+_34169036
|
0.397
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr9_+_34169058
|
0.394
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr11_-_66897584
|
0.355
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr9_+_34169047
|
0.354
|
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr17_-_76919068
|
0.354
|
NM_178520
|
TMEM105
|
transmembrane protein 105
|
|
chr10_+_70553983
|
0.349
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr17_+_35390632
|
0.348
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr9_+_34169002
|
0.343
|
NM_001171201
NM_001171202
NM_001171203
NM_001171204
NM_016525
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr6_+_117044334
|
0.335
|
NM_001010892
NM_001161664
|
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas)
|
|
chr6_-_134538762
|
0.334
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr8_-_29262240
|
0.328
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr5_-_64100176
|
0.316
|
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chr11_-_74120077
|
0.316
|
|
CHRDL2
|
chordin-like 2
|
|
chrX_+_505078
|
0.311
|
NM_000451
NM_006883
|
SHOX
|
short stature homeobox
|
|
chr11_-_8788765
|
0.311
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr17_+_35390641
|
0.310
|
|
PSMD3
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3
|
|
chr6_+_31698447
|
0.304
|
|
|
|
|
chr12_+_52060689
|
0.302
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr12_+_10222823
|
0.296
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr19_-_56164628
|
0.292
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr5_+_35892747
|
0.288
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr11_+_10433241
|
0.286
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr17_+_582406
|
0.282
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chr4_+_110956085
|
0.272
|
NM_018983
NM_032993
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr1_-_26070330
|
0.268
|
NM_178422
|
PAQR7
|
progestin and adipoQ receptor family member VII
|
|
chr5_-_64100239
|
0.265
|
NM_173829
|
SREK1IP1
|
SREK1-interacting protein 1
|
|
chrX_-_109448083
|
0.260
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr6_-_134538526
|
0.257
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_-_109447970
|
0.257
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chrX_-_62891712
|
0.254
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr11_-_8789457
|
0.248
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr1_-_224993498
|
0.243
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr18_-_44189625
|
0.241
|
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chrX_-_109447954
|
0.229
|
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr19_-_9098625
|
0.229
|
NM_001001958
|
OR7G3
|
olfactory receptor, family 7, subfamily G, member 3
|
|
chr9_-_83493415
|
0.225
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr12_+_83954229
|
0.222
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr6_-_143307976
|
0.220
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr19_-_53515143
|
0.220
|
NM_144577
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr3_-_129852345
|
0.219
|
|
RPN1
|
ribophorin I
|
|
chr4_+_120029426
|
0.217
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr3_-_193928038
|
0.216
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr1_-_20684858
|
0.213
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr1_+_151596953
|
0.212
|
NM_002965
|
S100A9
|
S100 calcium binding protein A9
|
|
chr1_-_20685314
|
0.210
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr10_+_70553949
|
0.209
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_70553862
|
0.208
|
NM_001035260
NM_004896
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_70553945
|
0.204
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr10_+_70553962
|
0.202
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr4_-_184662604
|
0.199
|
|
LOC389247
|
hypothetical LOC389247
|
|
chr10_+_70553936
|
0.198
|
|
VPS26A
|
vacuolar protein sorting 26 homolog A (S. pombe)
|
|
chr1_+_242691295
|
0.197
|
NM_001130957
NM_173807
|
C1orf101
|
chromosome 1 open reading frame 101
|
|
chrX_+_56606686
|
0.194
|
NM_013444
|
UBQLN2
|
ubiquilin 2
|
|
chr15_-_71712605
|
0.189
|
|
NPTN
|
neuroplastin
|
|
chr1_-_48235148
|
0.183
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr2_+_182464713
|
0.183
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr14_+_31616196
|
0.179
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr2_+_96269703
|
0.176
|
NM_001037228
|
LOC285033
|
hypothetical protein LOC285033
|
|
chr10_+_114699953
|
0.176
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_+_33130563
|
0.176
|
|
CRTAP
|
cartilage associated protein
|
|
chr4_+_84676280
|
0.175
|
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr13_+_56626195
|
0.174
|
NM_198441
NM_001130404
NM_001130405
NM_001130406
NM_001130407
|
PRR20A
PRR20B
PRR20C
PRR20D
PRR20E
|
proline rich 20A
proline rich 20B
proline rich 20C
proline rich 20D
proline rich 20E
|
|
chr17_-_60483346
|
0.173
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr22_+_38296703
|
0.169
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chr12_+_121803255
|
0.164
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr9_+_33807168
|
0.162
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr11_-_122571198
|
0.161
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr1_-_72520735
|
0.161
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr11_-_65244718
|
0.152
|
NM_032193
|
RNASEH2C
|
ribonuclease H2, subunit C
|
|
chr14_-_20636568
|
0.148
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chrX_-_49851743
|
0.146
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr17_-_70901303
|
0.144
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr12_-_40824699
|
0.143
|
NM_001099650
NM_173601
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr2_-_60634271
|
0.140
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr19_+_54786723
|
0.139
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr2_+_79593679
|
0.139
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr19_+_17972923
|
0.139
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr3_-_42282665
|
0.138
|
NM_000729
|
CCK
|
cholecystokinin
|
|
chr16_+_66438348
|
0.137
|
|
NUTF2
|
nuclear transport factor 2
|
|
chr15_-_56829225
|
0.135
|
NM_001110
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr3_-_15875932
|
0.135
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr10_-_103864579
|
0.135
|
|
LDB1
|
LIM domain binding 1
|
|
chr17_+_35372751
|
0.134
|
NM_178171
|
GSDMA
|
gasdermin A
|
|
chr11_+_10480891
|
0.134
|
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr7_+_21434818
|
0.131
|
|
|
|
|
chrX_+_122923279
|
0.128
|
|
STAG2
|
stromal antigen 2
|
|
chr4_+_84676414
|
0.127
|
NM_032717
|
AGPAT9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr14_+_69415880
|
0.127
|
NM_001034852
NM_022137
|
SMOC1
|
SPARC related modular calcium binding 1
|
|
chr9_-_103397023
|
0.123
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr12_+_29193399
|
0.122
|
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr10_-_14412813
|
0.119
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_-_27802688
|
0.118
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr9_+_33807213
|
0.116
|
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr12_-_92489652
|
0.115
|
|
LOC144481
|
hypothetical protein LOC144481
|
|
chr9_-_106705867
|
0.111
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr15_-_56829104
|
0.111
|
|
ADAM10
|
ADAM metallopeptidase domain 10
|
|
chr3_-_129852182
|
0.111
|
|
RPN1
|
ribophorin I
|
|
chr11_-_74119833
|
0.110
|
NM_015424
|
CHRDL2
|
chordin-like 2
|
|
chr15_-_86600658
|
0.109
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chrX_-_62891667
|
0.107
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr8_-_37943143
|
0.106
|
NM_000025
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr19_-_11234117
|
0.106
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr3_-_129852400
|
0.105
|
NM_002950
|
RPN1
|
ribophorin I
|
|
chrX_-_62891730
|
0.104
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr4_+_166519392
|
0.101
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr4_-_74343169
|
0.100
|
|
|
|
|
chr17_-_60483278
|
0.100
|
|
|
|
|
chr3_-_166397142
|
0.100
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr1_+_156229934
|
0.100
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr1_+_109036431
|
0.099
|
NM_018061
|
PRPF38B
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing B
|
|
chr19_-_47451119
|
0.098
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr3_-_121295663
|
0.098
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr3_-_188936941
|
0.096
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr2_+_79593567
|
0.096
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr4_+_110956360
|
0.094
|
|
GAR1
|
GAR1 ribonucleoprotein homolog (yeast)
|