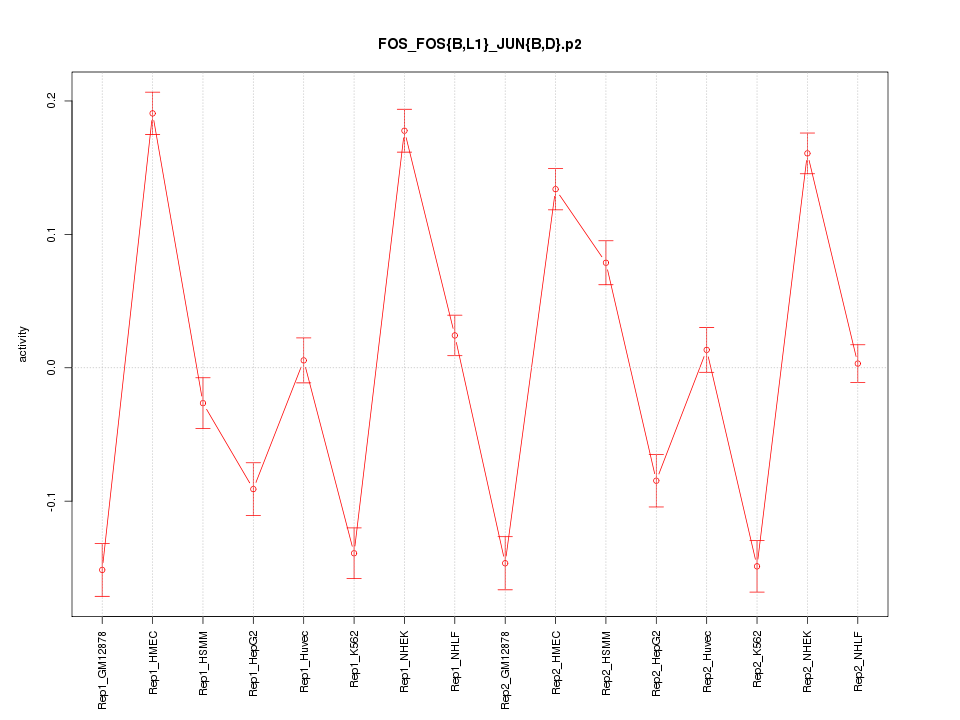
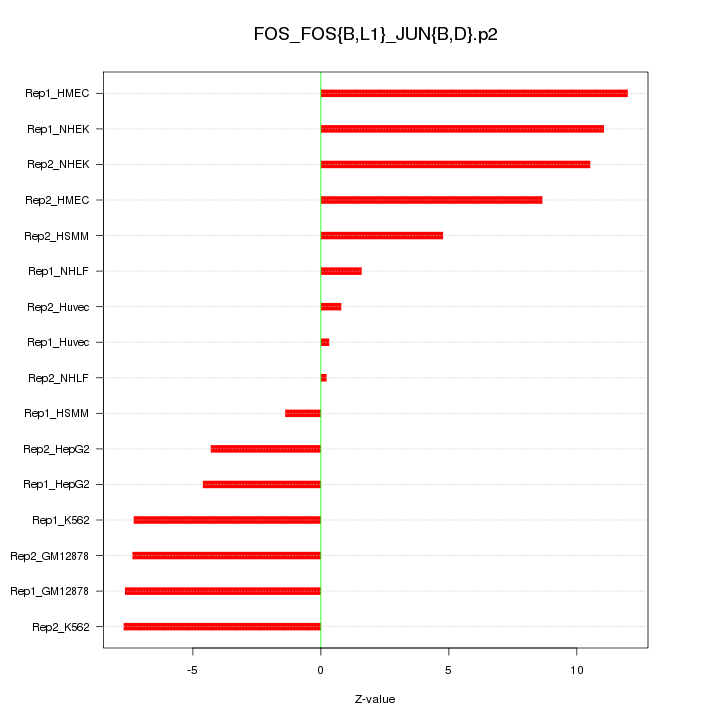
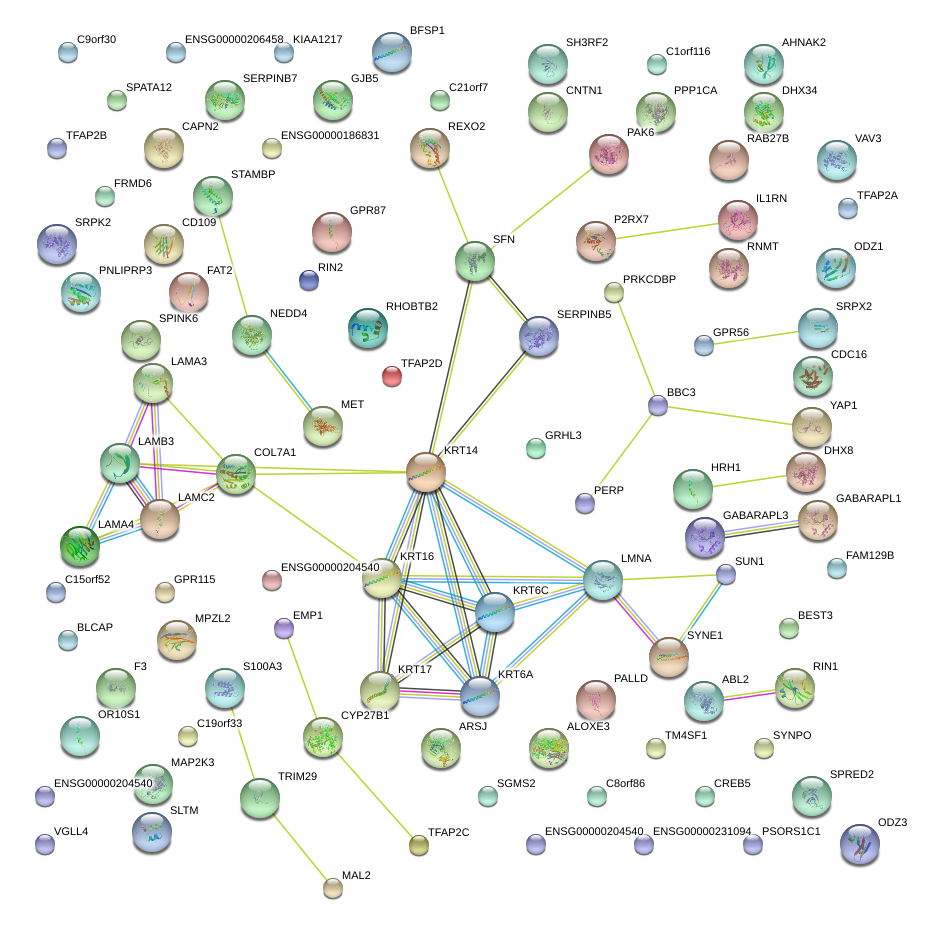
|
chr1_-_177378801
|
47.097
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr18_+_19706979
|
44.490
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr1_+_221955876
|
39.742
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr6_+_47774247
|
35.485
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr1_+_181421984
|
34.959
|
|
LAMC2
|
laminin, gamma 2
|
|
chr20_-_17487480
|
31.517
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr3_+_11153778
|
31.187
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr21_+_29439768
|
31.162
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr10_-_88720908
|
28.655
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr12_+_13240868
|
28.345
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240987
|
28.010
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240983
|
27.312
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr3_-_152517204
|
26.833
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr7_+_116099648
|
26.442
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr1_+_181421796
|
25.377
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr5_+_150000394
|
24.869
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr1_+_181422016
|
23.444
|
|
LAMC2
|
laminin, gamma 2
|
|
chr9_+_102231616
|
23.337
|
NM_001198806
|
C9orf30
|
chromosome 9 open reading frame 30
|
|
chr8_+_22900863
|
22.706
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr10_+_24795465
|
21.753
|
|
KIAA1217
|
KIAA1217
|
|
chr10_+_24795446
|
21.331
|
|
KIAA1217
|
KIAA1217
|
|
chr18_+_59593591
|
21.114
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr8_-_38505331
|
20.925
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr9_-_90456824
|
20.803
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr14_-_104507862
|
20.375
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr11_-_65860450
|
19.966
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr1_-_94779727
|
19.815
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr6_+_31190505
|
19.355
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr12_-_51173238
|
18.768
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr18_+_59295123
|
18.572
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr14_+_104218487
|
18.354
|
|
|
|
|
chr17_-_7962531
|
17.838
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr11_+_101488380
|
17.074
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_57069508
|
16.777
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr15_+_38319326
|
16.760
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr1_+_154351082
|
16.695
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr9_-_129381086
|
16.355
|
NM_001035534
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr17_-_36996611
|
16.325
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr4_+_108965167
|
16.007
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_145297490
|
15.974
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr1_-_207891264
|
15.572
|
NM_001017402
|
LAMB3
|
laminin, beta 3
|
|
chr1_+_34993234
|
15.334
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr20_+_19903778
|
15.174
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_-_68369322
|
14.812
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr1_-_36589744
|
14.754
|
|
|
|
|
chr1_-_108032645
|
14.560
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_+_27062215
|
14.425
|
NM_006142
|
SFN
|
stratifin
|
|
chr9_+_676640
|
13.972
|
|
|
|
|
chr17_+_21131940
|
13.847
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr10_+_118177371
|
13.828
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr11_-_6298284
|
13.710
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr7_+_838663
|
13.578
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr6_-_138470160
|
13.458
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr2_+_113591940
|
13.457
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr19_+_43486640
|
13.373
|
NM_033520
|
C19orf33
|
chromosome 19 open reading frame 33
|
|
chr6_-_152744143
|
13.325
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr16_+_56219802
|
13.252
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr12_+_10256682
|
13.094
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr1_-_205272619
|
13.006
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr6_-_152744022
|
12.919
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr3_-_150578103
|
12.911
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr4_-_115120251
|
12.905
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr4_+_183302105
|
12.872
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr11_-_119499188
|
12.836
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr2_-_65447267
|
12.817
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr3_-_11585264
|
12.603
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr1_+_154351200
|
12.552
|
|
LMNA
|
lamin A/C
|
|
chr19_-_52427698
|
12.417
|
NM_001127240
NM_001127241
NM_001127242
|
BBC3
|
BCL2 binding component 3
|
|
chr7_+_28692122
|
12.411
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr3_-_150578207
|
12.297
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_+_73911477
|
12.245
|
|
STAMBP
|
STAM binding protein
|
|
chr3_-_150578148
|
12.239
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr1_+_24518501
|
12.180
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_-_151788341
|
12.142
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr1_+_154351162
|
12.129
|
|
LMNA
|
lamin A/C
|
|
chr18_+_50646616
|
12.080
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr12_-_56445653
|
12.020
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr14_+_51025604
|
12.016
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr17_-_7962859
|
11.963
|
NM_001165960
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr3_-_150578023
|
11.807
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_-_65447370
|
11.513
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr15_-_38417564
|
11.334
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr6_-_10527772
|
11.247
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chrX_+_99786044
|
11.245
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr4_+_169989680
|
11.238
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr11_-_123353607
|
11.161
|
NM_001004474
|
OR10S1
|
olfactory receptor, family 10, subfamily S, member 1
|
|
chr20_-_35586366
|
11.161
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr11_-_117640166
|
11.128
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr7_-_104696678
|
11.087
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr17_-_37034288
|
11.054
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr1_+_24518398
|
10.768
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chrX_+_99785818
|
10.663
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr3_-_48607596
|
10.567
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr5_+_147562531
|
10.560
|
NM_001195290
NM_205841
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6
|
|
chr17_-_37022465
|
10.477
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr6_+_74462654
|
10.417
|
|
CD109
|
CD109 molecule
|
|
chr8_+_120289735
|
10.411
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr15_-_53996597
|
10.395
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr11_-_117640078
|
10.080
|
|
|
|
|
chr3_+_30623376
|
10.032
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr7_-_42243142
|
10.017
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr17_-_7434113
|
9.979
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr3_+_30622905
|
9.882
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr6_+_41712907
|
9.856
|
|
MDFI
|
MyoD family inhibitor
|
|
chr3_+_161877640
|
9.807
|
NM_025047
|
ARL14
|
ADP-ribosylation factor-like 14
|
|
chr11_+_12265022
|
9.751
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr18_+_54039818
|
9.692
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr21_-_35343065
|
9.604
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_-_117640196
|
9.570
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr20_-_1257782
|
9.544
|
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chrX_-_19598912
|
9.526
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr5_+_145296313
|
9.510
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr13_+_77007809
|
9.509
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr2_-_166359016
|
9.471
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr7_-_42243318
|
9.418
|
|
GLI3
|
GLI family zinc finger 3
|
|
chr18_+_54039779
|
9.411
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_73170290
|
9.262
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr6_+_74462226
|
9.215
|
NM_001159587
NM_001159588
NM_133493
|
CD109
|
CD109 molecule
|
|
chr11_+_35596310
|
9.172
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr11_-_117640407
|
9.148
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr20_-_1257809
|
8.991
|
NM_080489
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr6_+_74462485
|
8.991
|
|
CD109
|
CD109 molecule
|
|
chr18_+_54039730
|
8.984
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr19_-_43955984
|
8.854
|
NM_002307
|
LGALS7
LGALS7B
|
lectin, galactoside-binding, soluble, 7
lectin, galactoside-binding, soluble, 7B
|
|
chr6_-_116799548
|
8.806
|
|
LOC100287467
|
hypothetical LOC100287467
|
|
chr9_-_116920232
|
8.686
|
NM_002160
|
TNC
|
tenascin C
|
|
chr18_+_59788242
|
8.620
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr6_+_12398514
|
8.573
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr11_-_47502115
|
8.554
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr1_+_44171578
|
8.510
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr11_-_2907169
|
8.445
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr17_-_36928604
|
8.370
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr15_+_38318528
|
8.327
|
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr19_-_3920736
|
8.304
|
NM_001348
|
DAPK3
|
death-associated protein kinase 3
|
|
chr1_-_150276064
|
8.302
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr12_-_51912101
|
8.228
|
NM_000966
|
RARG
|
retinoic acid receptor, gamma
|
|
chr3_-_31997738
|
8.197
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr15_+_38849450
|
8.174
|
NM_001130448
|
C15orf62
|
chromosome 15 open reading frame 62
|
|
chr19_-_56215084
|
8.149
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr16_-_21221872
|
8.131
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr5_+_35892747
|
8.118
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr5_-_1935764
|
8.080
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr1_-_150276114
|
8.038
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr15_-_58477407
|
8.036
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr4_+_35959638
|
7.965
|
NM_001136536
|
DTHD1
|
death domain containing 1
|
|
chr18_+_59705918
|
7.951
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr17_-_36469869
|
7.943
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr21_-_35181956
|
7.920
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr11_-_33701073
|
7.846
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr20_+_33223434
|
7.823
|
NM_006404
|
PROCR
|
protein C receptor, endothelial
|
|
chr11_+_20005704
|
7.819
|
|
NAV2
|
neuron navigator 2
|
|
chr19_-_48866111
|
7.759
|
NM_001005376
NM_001005377
NM_002659
|
PLAUR
|
plasminogen activator, urokinase receptor
|
|
chr16_+_55199978
|
7.611
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr10_+_123862430
|
7.599
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr7_+_1543644
|
7.538
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_-_3407780
|
7.455
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr17_-_37196463
|
7.452
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr14_+_95792299
|
7.419
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr13_-_29321599
|
7.407
|
|
UBL3
|
ubiquitin-like 3
|
|
chr22_-_30388165
|
7.404
|
|
PISD
|
phosphatidylserine decarboxylase
|
|
chr1_+_28637247
|
7.309
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr14_+_62740878
|
7.290
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr6_-_112033775
|
7.265
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr17_-_24068759
|
7.258
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr15_+_65205107
|
7.230
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr3_+_48482213
|
7.158
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr5_+_73145098
|
7.149
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr1_-_151804929
|
7.130
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr11_-_8695974
|
7.090
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_-_112033752
|
7.082
|
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr3_-_123995216
|
6.947
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr5_+_36642424
|
6.854
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_-_118286822
|
6.817
|
NM_182557
|
BCL9L
|
B-cell CLL/lymphoma 9-like
|
|
chr6_+_74462630
|
6.792
|
|
CD109
|
CD109 molecule
|
|
chr11_-_66898223
|
6.746
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr17_+_71229110
|
6.707
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr9_-_109291866
|
6.696
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_15706340
|
6.673
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr5_-_148422799
|
6.670
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr3_-_31998241
|
6.643
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr3_+_31998269
|
6.627
|
NM_001137674
|
ZNF860
|
zinc finger protein 860
|
|
chr1_-_89261021
|
6.552
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr14_+_20537253
|
6.534
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr11_+_58146721
|
6.519
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr17_-_3408038
|
6.503
|
NM_145068
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr7_-_16811131
|
6.469
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr1_+_40508336
|
6.466
|
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr1_+_35019376
|
6.359
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr4_-_152366970
|
6.351
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr9_-_34652629
|
6.343
|
NM_006664
|
CCL27
|
chemokine (C-C motif) ligand 27
|
|
chr3_-_127258299
|
6.316
|
NM_001008487
|
SLC41A3
|
solute carrier family 41, member 3
|
|
chr17_+_71229151
|
6.291
|
|
ITGB4
|
integrin, beta 4
|
|
chr6_+_83129641
|
6.278
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr15_+_38923470
|
6.273
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr18_-_45630154
|
6.266
|
|
MYO5B
|
myosin VB
|
|
chr3_+_69216747
|
6.263
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr18_-_26996742
|
6.199
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr6_+_1556543
|
6.196
|
|
FOXC1
|
forkhead box C1
|
|
chr6_-_35804337
|
6.109
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr11_-_101906640
|
6.106
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|