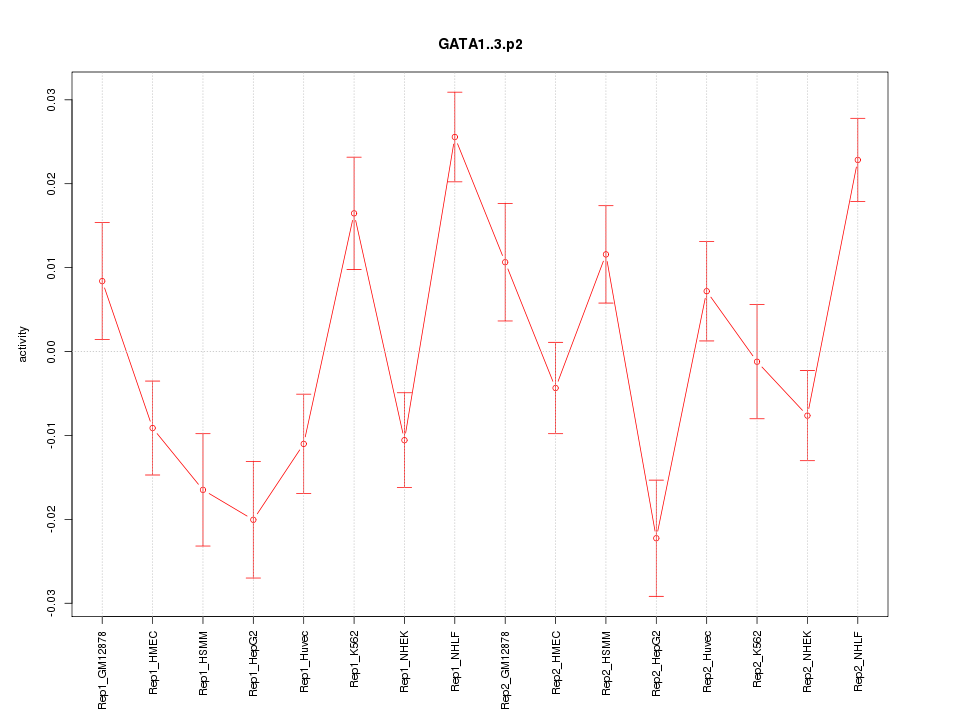
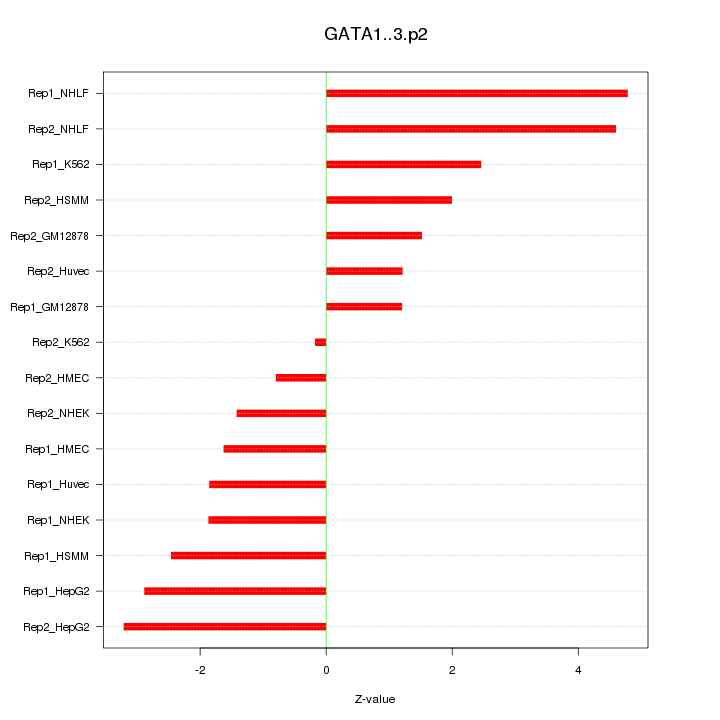
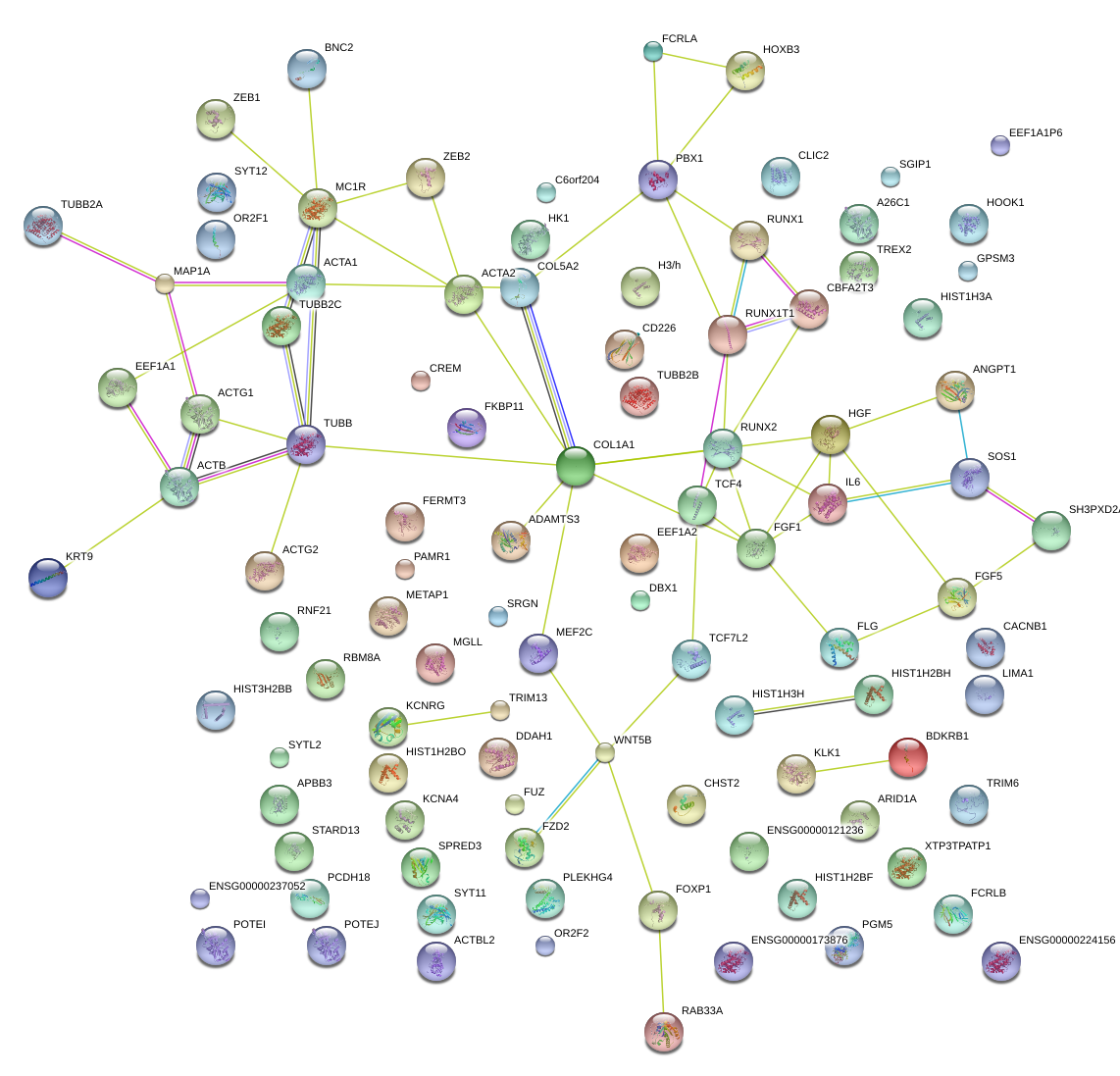
|
chr17_+_39990124
|
2.602
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr6_-_74284522
|
2.401
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr8_-_108579270
|
2.392
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr11_-_85107569
|
2.316
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_-_16717887
|
2.184
|
|
BNC2
|
basonuclin 2
|
|
chr15_+_41597097
|
2.128
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr9_+_70161634
|
2.112
|
NM_021965
|
PGM5
|
phosphoglucomutase 5
|
|
chr6_-_119137923
|
2.038
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr1_-_85816520
|
1.880
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_+_140871107
|
1.800
|
|
|
|
|
chr1_+_66772393
|
1.797
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr7_+_143263191
|
1.775
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chr2_-_189752575
|
1.753
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr12_-_48902692
|
1.681
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr17_-_45629401
|
1.641
|
|
|
|
|
chr7_-_5534252
|
1.610
|
|
ACTB
|
actin, beta
|
|
chr5_+_140870881
|
1.583
|
|
|
|
|
chr5_-_141973875
|
1.564
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_-_93099040
|
1.487
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr10_+_35524060
|
1.478
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_30799130
|
1.474
|
|
TUBB
|
tubulin, beta
|
|
chr4_-_138673078
|
1.463
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chrX_-_154216929
|
1.452
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr17_-_45626364
|
1.423
|
|
|
|
|
chr10_-_105427898
|
1.422
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_73653331
|
1.412
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr16_-_87570716
|
1.396
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr2_-_144991386
|
1.370
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_85108030
|
1.363
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_-_88214720
|
1.356
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_+_43572679
|
1.342
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr17_-_45633873
|
1.334
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr7_-_81237162
|
1.324
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr1_+_162795538
|
1.298
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr10_+_70517863
|
1.283
|
|
SRGN
|
serglycin
|
|
chr2_-_144994349
|
1.278
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_144991556
|
1.260
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_35503720
|
1.251
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr10_+_70517823
|
1.251
|
NM_002727
|
SRGN
|
serglycin
|
|
chr6_-_32268267
|
1.251
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr6_-_26379567
|
1.216
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chrX_+_129133234
|
1.211
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr1_+_162795328
|
1.210
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr7_-_5534737
|
1.190
|
|
ACTB
|
actin, beta
|
|
chr18_-_51219880
|
1.173
|
|
TCF4
|
transcription factor 4
|
|
chr14_+_95792299
|
1.162
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr13_-_32678142
|
1.153
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_-_138673014
|
1.151
|
|
PCDH18
|
protocadherin 18
|
|
chr2_-_144994038
|
1.142
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_129023850
|
1.125
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr19_-_55008172
|
1.112
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr3_-_71262426
|
1.102
|
|
FOXP1
|
forkhead box P1
|
|
chr10_+_70745615
|
1.095
|
NM_033496
|
HK1
|
hexokinase 1
|
|
chr1_+_26973804
|
1.088
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr16_+_65868913
|
1.073
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr21_-_35153664
|
1.065
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_159959072
|
1.057
|
NM_001002901
|
FCRLB
|
Fc receptor-like B
|
|
chr1_+_226712430
|
1.047
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr1_-_32479785
|
1.044
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr11_+_63730720
|
1.042
|
NM_031471
NM_178443
|
FERMT3
|
fermitin family member 3
|
|
chr4_+_81406765
|
1.011
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr1_+_154095883
|
0.997
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr1_+_144218960
|
0.991
|
NM_005105
|
RBM8A
|
RNA binding motif protein 8A
|
|
chr5_-_139924229
|
0.990
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr17_-_44022595
|
0.976
|
|
HOXB3
|
homeobox B3
|
|
chr3_-_71262405
|
0.975
|
|
FOXP1
|
forkhead box P1
|
|
chr11_+_5610060
|
0.973
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr13_+_49487390
|
0.964
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr12_+_1608672
|
0.963
|
NM_032642
|
WNT5B
|
wingless-type MMTV integration site family, member 5B
|
|
chr18_-_65766068
|
0.961
|
|
CD226
|
CD226 molecule
|
|
chr3_+_144321286
|
0.960
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr3_+_142587973
|
0.960
|
|
|
|
|
chr6_+_33151703
|
0.952
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr4_-_89837951
|
0.950
|
NM_153757
|
NAP1L5
|
nucleosome assembly protein 1-like 5
|
|
chr13_-_107317460
|
0.942
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr20_-_49612745
|
0.937
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr6_-_26393705
|
0.934
|
NM_003543
|
HIST1H4H
|
histone cluster 1, H4h
|
|
chr17_+_35853177
|
0.931
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr15_-_41589958
|
0.930
|
NM_005657
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr11_+_62136788
|
0.930
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr3_-_11598829
|
0.927
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr2_-_38156811
|
0.926
|
NM_000104
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr16_+_29582480
|
0.922
|
|
SPN
|
sialophorin
|
|
chr2_-_38156769
|
0.919
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr11_-_47356479
|
0.913
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr1_-_26517342
|
0.912
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr2_+_55600220
|
0.912
|
NM_080667
|
CCDC104
|
coiled-coil domain containing 104
|
|
chr2_+_157822355
|
0.909
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr8_-_93144039
|
0.907
|
|
|
|
|
chr7_+_50318801
|
0.905
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr4_+_72423633
|
0.895
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_51208499
|
0.885
|
|
CDKN2C
|
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4)
|
|
chr11_+_63730766
|
0.881
|
|
FERMT3
|
fermitin family member 3
|
|
chr5_+_176494690
|
0.880
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr15_+_55298908
|
0.877
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr10_-_105202139
|
0.871
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr19_+_1221925
|
0.870
|
|
CIRBP
|
cold inducible RNA binding protein
|
|
chr6_+_26381122
|
0.868
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr4_+_160408447
|
0.862
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr1_-_226712140
|
0.855
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr11_-_47356562
|
0.845
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr10_-_105202012
|
0.840
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr17_-_44026042
|
0.833
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr15_-_75259714
|
0.813
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr5_-_158459276
|
0.813
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr6_+_26128656
|
0.809
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr7_-_13996166
|
0.804
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr6_-_29120930
|
0.798
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chr19_-_59383944
|
0.794
|
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7
|
|
chr3_-_49826221
|
0.783
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chrX_-_100527799
|
0.782
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr2_-_74634569
|
0.775
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr11_-_64403712
|
0.775
|
|
EHD1
|
EH-domain containing 1
|
|
chr10_-_13789628
|
0.768
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr13_-_26643775
|
0.762
|
|
USP12
|
ubiquitin specific peptidase 12
|
|
chr5_+_139924224
|
0.760
|
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr1_-_72521079
|
0.760
|
|
|
|
|
chr1_-_153509473
|
0.747
|
NM_003993
|
CLK2
|
CDC-like kinase 2
|
|
chr17_-_53761119
|
0.739
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr17_-_77092569
|
0.732
|
|
ACTG1
|
actin, gamma 1
|
|
chr2_+_202607554
|
0.731
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr9_-_122728173
|
0.726
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr20_+_19903778
|
0.724
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr7_+_90176647
|
0.719
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr1_+_81544432
|
0.718
|
|
LPHN2
|
latrophilin 2
|
|
chr3_-_145049853
|
0.716
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr2_-_175337386
|
0.715
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr1_+_31655607
|
0.713
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr1_+_114323552
|
0.710
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr3_+_120496180
|
0.702
|
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr1_-_200182090
|
0.692
|
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr15_+_45797977
|
0.683
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr1_-_153209625
|
0.682
|
NM_001130040
NM_183001
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr3_-_33675484
|
0.680
|
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr10_-_80875097
|
0.678
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr15_-_78050515
|
0.678
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr3_-_55489973
|
0.676
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr17_-_44026321
|
0.672
|
|
HOXB5
|
homeobox B5
|
|
chr1_-_200182338
|
0.671
|
NM_012134
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr6_-_32268615
|
0.670
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr7_+_86619580
|
0.668
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr1_+_154120170
|
0.667
|
|
SYT11
|
synaptotagmin XI
|
|
chr1_-_148051859
|
0.667
|
NM_001123375
|
HIST2H3D
|
histone cluster 2, H3d
|
|
chr5_-_81082699
|
0.665
|
NM_012446
|
SSBP2
|
single-stranded DNA binding protein 2
|
|
chr4_+_89147821
|
0.663
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr12_+_46799279
|
0.662
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr3_-_49826360
|
0.662
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr13_-_26643952
|
0.661
|
NM_182488
|
USP12
|
ubiquitin specific peptidase 12
|
|
chrX_-_57953578
|
0.655
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr22_+_20715340
|
0.655
|
|
|
|
|
chr1_+_234372454
|
0.654
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr12_-_55293131
|
0.650
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr4_+_141397889
|
0.647
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr8_+_70567634
|
0.646
|
|
SULF1
|
sulfatase 1
|
|
chr22_-_35970133
|
0.644
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr9_+_133154896
|
0.644
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr8_-_93144366
|
0.639
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_47356650
|
0.639
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr2_+_170074457
|
0.636
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr6_+_44076281
|
0.632
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr10_+_104995633
|
0.632
|
NM_001143909
|
LOC729020
|
rcRPE
|
|
chr6_-_76259976
|
0.632
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr15_-_41185460
|
0.631
|
NM_174916
|
UBR1
|
ubiquitin protein ligase E3 component n-recognin 1
|
|
chr6_-_32203971
|
0.628
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr17_-_43977273
|
0.625
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr11_-_62233552
|
0.625
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr14_+_54291255
|
0.624
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr4_+_54790190
|
0.623
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr9_+_18464138
|
0.622
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr10_-_75080784
|
0.621
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr16_+_29581783
|
0.618
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr1_+_152566899
|
0.617
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr11_-_47356672
|
0.616
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr19_-_45888259
|
0.615
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr5_-_1547872
|
0.612
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr16_+_11966464
|
0.611
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr6_-_130224108
|
0.607
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr6_-_32203952
|
0.605
|
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr11_-_47356642
|
0.596
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr22_-_37598115
|
0.593
|
|
CBX6
|
chromobox homolog 6
|
|
chr12_-_55295628
|
0.586
|
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr22_-_43272100
|
0.585
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr17_-_45626967
|
0.582
|
|
|
|
|
chr8_+_70567581
|
0.580
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr9_+_112470871
|
0.578
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr7_+_130445394
|
0.578
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr5_-_178977441
|
0.577
|
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H)
|
|
chr3_+_49002285
|
0.575
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr22_-_37598160
|
0.574
|
|
CBX6
|
chromobox homolog 6
|
|
chr6_-_30763057
|
0.574
|
NM_001134870
|
KIAA1949
|
KIAA1949
|
|
chr1_+_153232685
|
0.570
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr17_-_44641741
|
0.570
|
NM_031498
NM_001198755
NM_001198756
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr1_-_72520735
|
0.570
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr5_-_71838875
|
0.568
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr1_+_207944807
|
0.568
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr12_-_113330574
|
0.567
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr17_-_38229790
|
0.567
|
NM_003766
|
BECN1
|
beclin 1, autophagy related
|
|
chr1_+_246682699
|
0.565
|
NM_001004136
|
OR2T2
|
olfactory receptor, family 2, subfamily T, member 2
|
|
chr22_-_35875890
|
0.564
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr11_+_4859624
|
0.562
|
NM_001004759
|
OR51T1
|
olfactory receptor, family 51, subfamily T, member 1
|