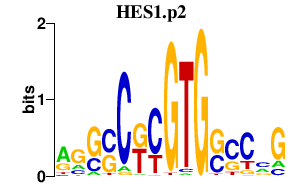
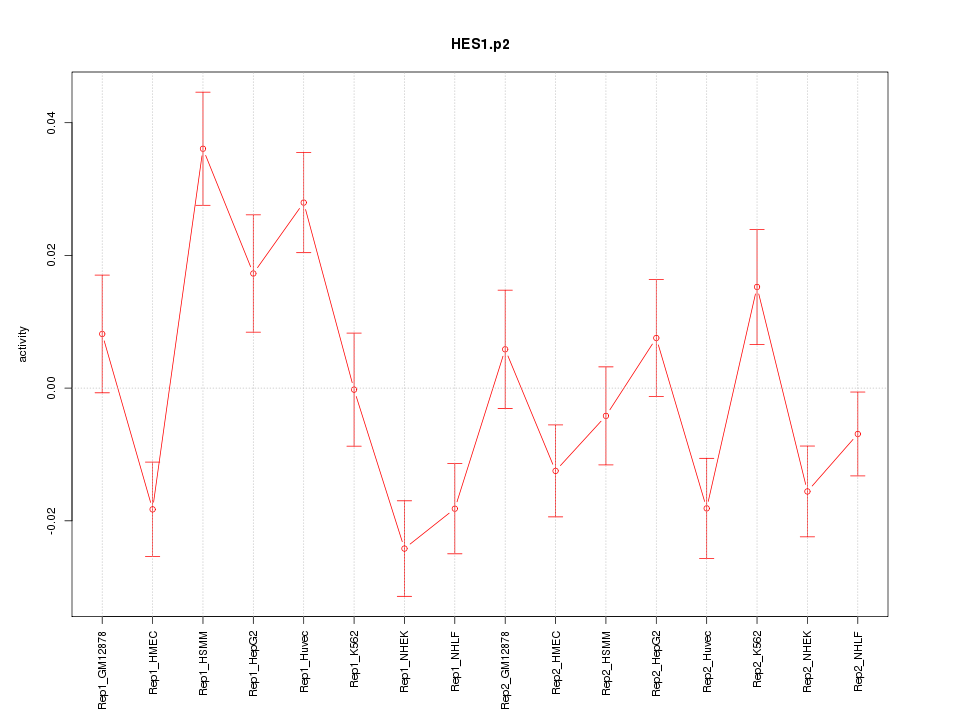
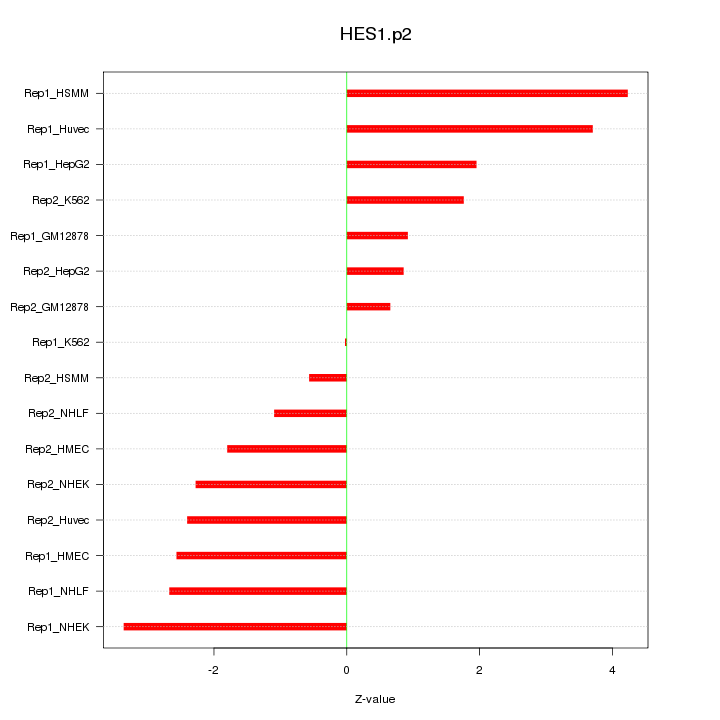
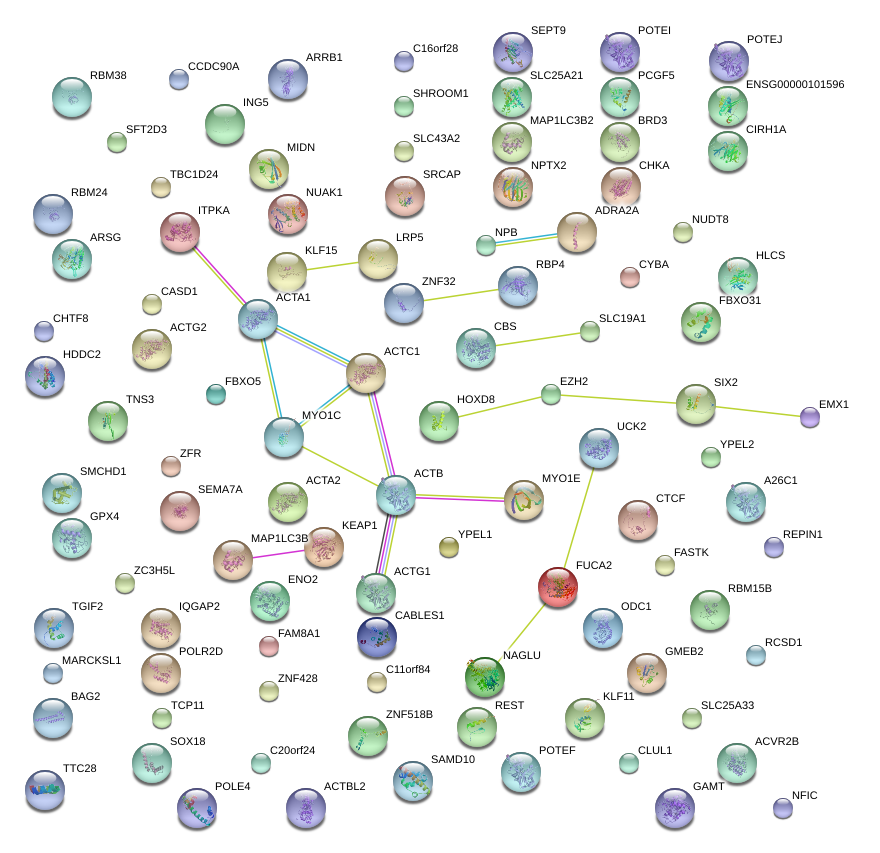
|
chr18_+_18969724
|
3.279
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr16_+_66153810
|
1.836
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr21_-_45786709
|
1.636
|
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr3_+_129690735
|
1.605
|
|
|
|
|
chr22_-_27405708
|
1.535
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr17_+_37941715
|
1.501
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr10_+_92912676
|
1.409
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr19_-_10474767
|
1.399
|
NM_203500
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr17_+_37941790
|
1.340
|
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr19_-_10475083
|
1.329
|
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr7_+_98084531
|
1.323
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr17_+_37941476
|
1.279
|
NM_000263
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr3_+_51403760
|
1.264
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr7_+_149696263
|
1.262
|
|
REPIN1
|
replication initiator 1
|
|
chr2_-_10506033
|
1.262
|
|
ODC1
|
ornithine decarboxylase 1
|
|
chr5_+_75734759
|
1.256
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr2_+_10101822
|
1.238
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr2_-_10505902
|
1.168
|
NM_002539
|
ODC1
|
ornithine decarboxylase 1
|
|
chr11_-_67645246
|
1.166
|
|
CHKA
|
choline kinase alpha
|
|
chr12_+_6893751
|
1.160
|
NM_001975
|
ENO2
|
enolase 2 (gamma, neuronal)
|
|
chr2_+_242289983
|
1.160
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr10_+_92970487
|
1.150
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr10_+_92970302
|
1.148
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr17_-_77094426
|
1.118
|
|
ACTG1
|
actin, gamma 1
|
|
chr11_-_67153959
|
1.109
|
NM_181843
|
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8
|
|
chr19_+_1199548
|
1.094
|
NM_177401
|
MIDN
|
midnolin
|
|
chr19_-_10474266
|
1.075
|
NM_012289
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr2_+_75039273
|
1.061
|
NM_019896
|
POLE4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit)
|
|
chr9_-_135922912
|
1.057
|
NM_007371
|
BRD3
|
bromodomain containing 3
|
|
chr7_+_149696792
|
1.053
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr6_-_125664913
|
1.026
|
NM_016063
|
HDDC2
|
HD domain containing 2
|
|
chr11_+_67836575
|
1.016
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr16_+_30617962
|
1.015
|
NM_006662
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr7_-_5536704
|
1.014
|
NM_001101
|
ACTB
|
actin, beta
|
|
chr19_-_1352474
|
1.014
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr20_-_62151216
|
1.010
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr20_-_61728866
|
1.002
|
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr6_-_13922525
|
0.995
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr7_-_23476156
|
0.994
|
|
|
|
|
chr16_-_67723971
|
0.985
|
|
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae)
|
|
chr1_+_9522107
|
0.984
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr6_-_163754903
|
0.981
|
|
LOC100526820
|
hypothetical LOC100526820
|
|
chr2_+_128175413
|
0.963
|
|
SFT2D3
|
SFT2 domain containing 3
|
|
chr7_-_47588266
|
0.951
|
|
TNS3
|
tensin 3
|
|
chr19_-_48815853
|
0.945
|
NM_182498
|
ZNF428
|
zinc finger protein 428
|
|
chr16_-_87244896
|
0.943
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr7_-_47588160
|
0.940
|
|
TNS3
|
tensin 3
|
|
chr3_+_38470782
|
0.930
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr7_+_93977267
|
0.913
|
|
CASD1
|
CAS1 domain containing 1
|
|
chr7_+_149696901
|
0.906
|
|
REPIN1
|
replication initiator 1
|
|
chr5_-_32480486
|
0.899
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr4_-_10068055
|
0.896
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chr11_-_74740298
|
0.896
|
NM_004041
NM_020251
|
ARRB1
|
arrestin, beta 1
|
|
chr6_+_57145292
|
0.895
|
|
BAG2
|
BCL2-associated athanogene 2
|
|
chr1_+_9522133
|
0.893
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr1_-_32574185
|
0.889
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr21_-_43368963
|
0.888
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr7_-_150408785
|
0.885
|
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr17_-_77094375
|
0.884
|
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr1_+_165865953
|
0.883
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr2_-_45090025
|
0.879
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr6_+_17708483
|
0.878
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr12_-_105057940
|
0.858
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr4_+_57469122
|
0.856
|
|
REST
|
RE1-silencing transcription factor
|
|
chr15_+_39573363
|
0.855
|
NM_002220
|
ITPKA
|
inositol 1,4,5-trisphosphate 3-kinase A
|
|
chr6_-_143874463
|
0.854
|
NM_032020
|
FUCA2
|
fucosidase, alpha-L- 2, plasma
|
|
chr20_+_55399847
|
0.853
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr6_-_35216981
|
0.852
|
NM_001093728
NM_018679
|
TCP11
|
t-complex 11 homolog (mouse)
|
|
chr7_-_148212310
|
0.848
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_-_148212336
|
0.844
|
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr7_-_148212312
|
0.836
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr19_+_1055614
|
0.833
|
NM_001039848
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chr20_+_34635362
|
0.825
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr5_-_32480586
|
0.824
|
NM_016107
|
ZFR
|
zinc finger RNA binding protein
|
|
chr7_-_148212230
|
0.821
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr20_-_34635091
|
0.817
|
|
|
|
|
chr17_+_63799228
|
0.809
|
|
ARSG
|
arylsulfatase G
|
|
chr17_-_1341599
|
0.809
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr16_+_30015243
|
0.801
|
|
|
|
|
chr10_-_95350949
|
0.800
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr16_-_85974810
|
0.793
|
NM_024735
|
FBXO31
|
F-box protein 31
|
|
chr20_-_62081434
|
0.790
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr22_-_20419975
|
0.787
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr16_+_85982884
|
0.785
|
|
MAP1LC3B
|
microtubule-associated protein 1 light chain 3 beta
|
|
chr16_+_2465109
|
0.785
|
NM_020705
|
TBC1D24
|
TBC1 domain family, member 24
|
|
chr3_-_127558747
|
0.783
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr20_+_34667624
|
0.775
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr5_+_95093267
|
0.772
|
|
|
|
|
chr15_-_72513311
|
0.771
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr16_-_1369613
|
0.767
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr18_+_2645885
|
0.762
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr11_+_63337498
|
0.762
|
NM_138471
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr4_+_57469907
|
0.757
|
|
REST
|
RE1-silencing transcription factor
|
|
chr19_+_3310138
|
0.753
|
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr17_+_72880831
|
0.753
|
NM_001113493
|
SEPT9
|
septin 9
|
|
chr10_-_43464157
|
0.747
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr17_-_1478859
|
0.746
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr2_+_176702667
|
0.740
|
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr18_+_586997
|
0.738
|
NM_014410
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr5_-_132194488
|
0.734
|
NM_001172700
|
SHROOM1
|
shroom family member 1
|
|
chr16_+_67723988
|
0.733
|
NM_032830
|
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin)
|
|
chr21_-_37284351
|
0.732
|
NM_000411
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr1_+_164063276
|
0.729
|
NM_012474
|
UCK2
|
uridine-cytidine kinase 2
|
|
chr12_-_31635089
|
0.726
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr16_+_15397120
|
0.725
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr16_-_4406608
|
0.725
|
|
CORO7
|
coronin 7
|
|
chr5_+_176806401
|
0.724
|
NM_001174101
NM_030567
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr20_-_61728774
|
0.723
|
NM_012384
|
GMEB2
|
glucocorticoid modulatory element binding protein 2
|
|
chr17_-_73636362
|
0.723
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr16_-_4406596
|
0.720
|
|
CORO7
|
coronin 7
|
|
chr6_-_125664770
|
0.720
|
|
HDDC2
|
HD domain containing 2
|
|
chr16_+_67724023
|
0.719
|
|
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin)
|
|
chr20_-_31725821
|
0.719
|
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr12_+_2939250
|
0.713
|
|
TEAD4
|
TEA domain family member 4
|
|
chr9_+_130504782
|
0.708
|
|
PKN3
|
protein kinase N3
|
|
chr17_+_43374021
|
0.706
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr20_-_61317891
|
0.704
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr3_-_129690056
|
0.703
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr17_-_73636386
|
0.701
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr2_-_242096673
|
0.694
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr20_+_30871357
|
0.692
|
NM_012325
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr19_+_748448
|
0.690
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr19_-_44032202
|
0.688
|
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr19_+_34994701
|
0.684
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr9_-_85761451
|
0.681
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr16_-_960982
|
0.681
|
NM_022773
|
LMF1
|
lipase maturation factor 1
|
|
chr16_-_4604895
|
0.680
|
NM_145253
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr22_-_37481925
|
0.680
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr14_+_102313566
|
0.680
|
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr16_+_387176
|
0.680
|
NM_005009
|
NME4
|
non-metastatic cells 4, protein expressed in
|
|
chr5_-_132141368
|
0.679
|
NM_001098813
|
SEPT8
|
septin 8
|
|
chr2_+_223244606
|
0.678
|
NM_058165
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1
|
|
chr7_-_150408824
|
0.676
|
NM_006712
NM_033015
|
FASTK
|
Fas-activated serine/threonine kinase
|
|
chr20_+_30871438
|
0.675
|
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1
|
|
chr20_-_31725915
|
0.672
|
NM_031231
NM_031232
|
NECAB3
|
N-terminal EF-hand calcium binding protein 3
|
|
chr13_-_19997914
|
0.669
|
NM_015974
|
CRYL1
|
crystallin, lambda 1
|
|
chr19_+_54309423
|
0.665
|
NM_022165
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr17_-_77422526
|
0.665
|
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chr5_+_95092384
|
0.661
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr11_-_67645433
|
0.660
|
NM_001277
NM_212469
|
CHKA
|
choline kinase alpha
|
|
chr16_-_4604781
|
0.659
|
|
FAM100A
|
family with sequence similarity 100, member A
|
|
chr16_-_65775326
|
0.658
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr20_-_7948370
|
0.657
|
NM_021156
|
TMX4
|
thioredoxin-related transmembrane protein 4
|
|
chr7_+_93976927
|
0.651
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr9_-_85761472
|
0.651
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr19_+_35124951
|
0.646
|
NM_003796
NM_134447
|
C19orf2
|
chromosome 19 open reading frame 2
|
|
chr20_+_34667605
|
0.643
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr17_-_3546108
|
0.643
|
|
P2RX5-TAX1BP3
P2RX5
|
P2RX5-TAX1BP3 read-through transcript
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr7_-_45927263
|
0.642
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr2_+_61146866
|
0.639
|
NM_032506
|
KIAA1841
|
KIAA1841
|
|
chr8_-_11242408
|
0.639
|
|
LOC100129129
|
hypothetical LOC100129129
|
|
chr22_-_22440054
|
0.638
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr1_+_36046373
|
0.638
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr16_-_88084387
|
0.635
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr1_-_1700088
|
0.627
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr19_+_748452
|
0.627
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr16_-_4406571
|
0.625
|
|
CORO7-PAM16
CORO7
|
CORO7-PAM16 read-through transcript
coronin 7
|
|
chr21_+_44377913
|
0.625
|
NM_004649
NM_198155
|
C21orf33
|
chromosome 21 open reading frame 33
|
|
chr15_+_83160914
|
0.623
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr22_-_37481403
|
0.623
|
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr19_+_54309443
|
0.622
|
|
LIN7B
|
lin-7 homolog B (C. elegans)
|
|
chr20_+_34667597
|
0.621
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr5_-_133368288
|
0.621
|
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr16_+_2742677
|
0.618
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr11_+_63337421
|
0.614
|
|
C11orf84
|
chromosome 11 open reading frame 84
|
|
chr16_-_65774904
|
0.613
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr22_-_37481857
|
0.612
|
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr1_-_233879617
|
0.612
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr5_-_172594691
|
0.612
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr22_-_49311439
|
0.609
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr16_+_83291055
|
0.608
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr6_+_31976754
|
0.605
|
NM_001178063
|
C2
|
complement component 2
|
|
chr8_-_12657339
|
0.603
|
NM_152271
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chrX_-_18282698
|
0.602
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr19_+_59385930
|
0.601
|
NM_024075
|
TSEN34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae)
|
|
chr7_-_149651594
|
0.600
|
NM_001164458
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast)
|
|
chr4_-_2233606
|
0.597
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr2_+_10360471
|
0.597
|
NM_002149
|
HPCAL1
|
hippocalcin-like 1
|
|
chr5_-_1547872
|
0.597
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr7_+_149651202
|
0.595
|
NM_001142928
NM_023942
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr17_+_43373887
|
0.594
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr21_-_39642880
|
0.594
|
NM_004965
|
HMGN1
|
high-mobility group nucleosome binding domain 1
|
|
chr20_+_34667566
|
0.592
|
NM_018840
NM_199483
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr9_+_36126710
|
0.591
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr15_+_83326317
|
0.591
|
|
PDE8A
|
phosphodiesterase 8A
|
|
chr17_-_77422561
|
0.589
|
NM_001185078
NM_004309
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha
|
|
chrX_-_7076146
|
0.588
|
NM_001135565
NM_001178135
NM_001178136
NM_012080
|
HDHD1
|
haloacid dehalogenase-like hydrolase domain containing 1
|
|
chr12_+_100615530
|
0.588
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr19_+_55045754
|
0.584
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr6_+_89912464
|
0.583
|
NM_001010853
|
PM20D2
|
peptidase M20 domain containing 2
|
|
chr7_+_149651481
|
0.583
|
|
LRRC61
|
leucine rich repeat containing 61
|
|
chr20_+_34667590
|
0.577
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr17_-_77411803
|
0.577
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr9_+_36126667
|
0.576
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr2_-_236741352
|
0.576
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr7_+_142205266
|
0.575
|
|
|
|
|
chr14_+_99507848
|
0.574
|
|
EVL
|
Enah/Vasp-like
|
|
chr17_-_77488258
|
0.574
|
NM_006907
NM_153824
|
PYCR1
|
pyrroline-5-carboxylate reductase 1
|
|
chr17_-_59131210
|
0.573
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr4_+_57468792
|
0.571
|
NM_005612
|
REST
|
RE1-silencing transcription factor
|