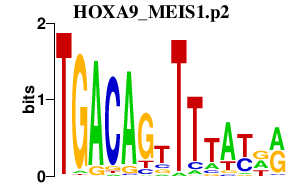
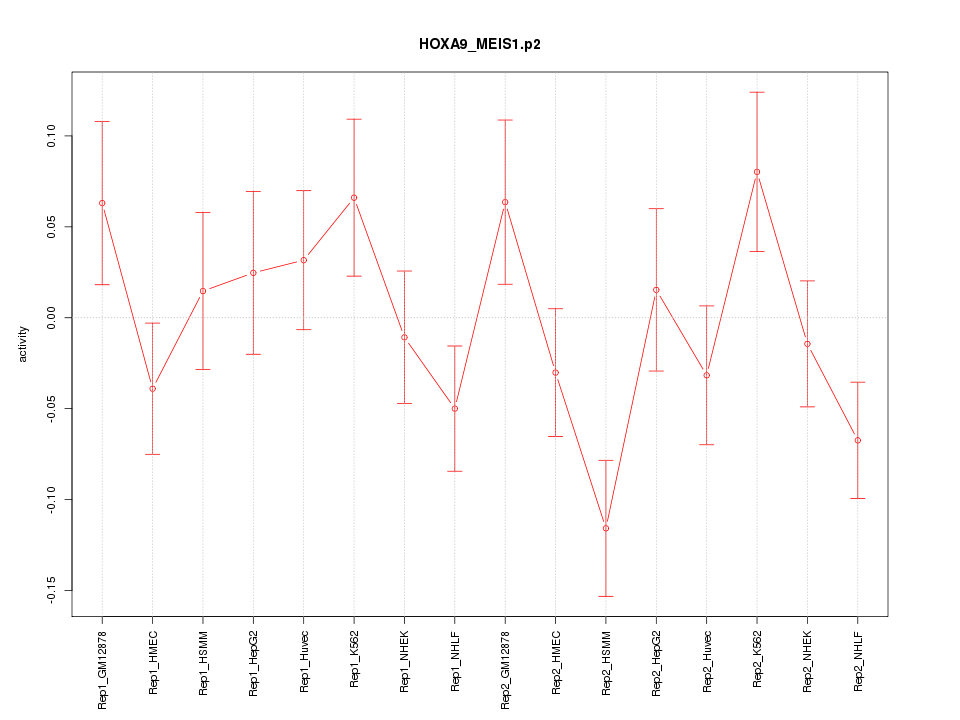
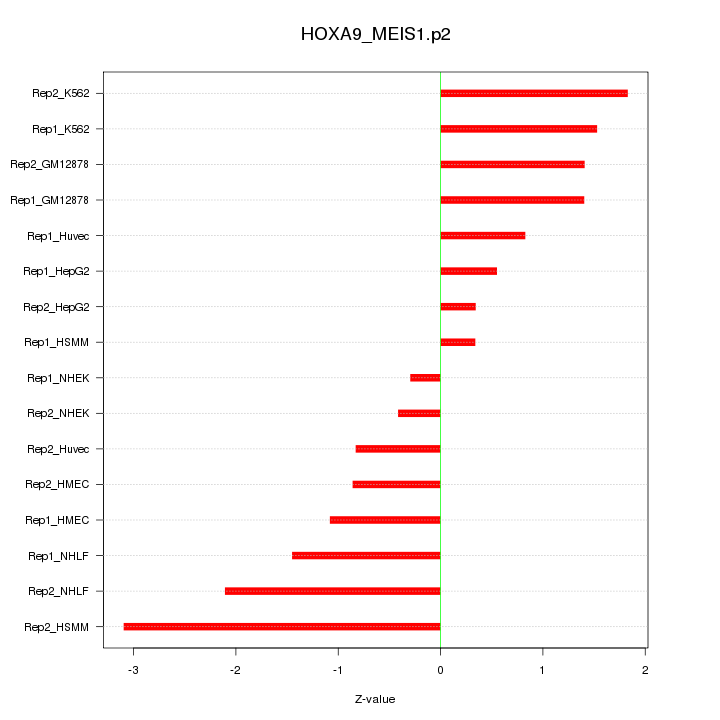
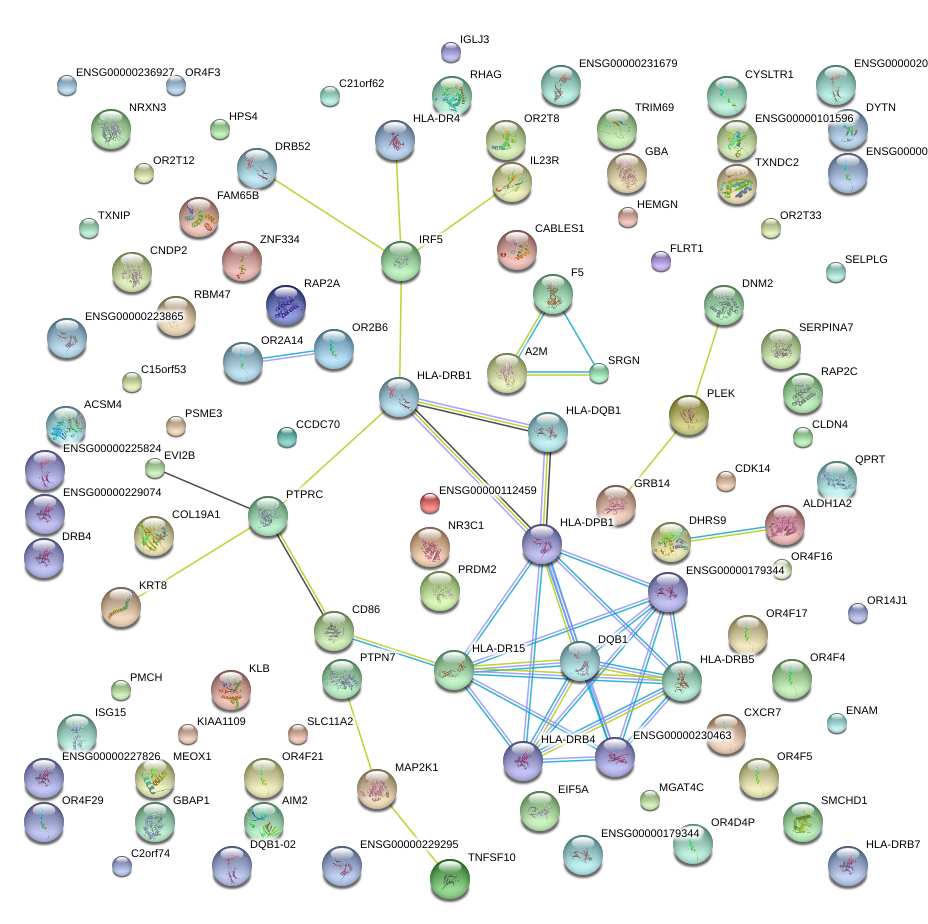
|
chr18_+_9875722
|
3.740
|
NM_001098529
NM_032243
|
TXNDC2
|
thioredoxin domain containing 2 (spermatozoa)
|
|
chr16_+_29597965
|
3.568
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr6_-_25044156
|
2.507
|
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr15_+_42815851
|
2.243
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr16_+_29597829
|
2.171
|
NM_014298
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr1_+_196874839
|
2.153
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_-_611896
|
2.091
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr9_-_99740243
|
1.911
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr2_+_68445821
|
1.737
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr1_-_157313179
|
1.416
|
NM_004833
|
AIM2
|
absent in melanoma 2
|
|
chr4_-_40211324
|
1.398
|
|
RBM47
|
RNA binding motif protein 47
|
|
chrX_-_130792297
|
1.355
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr10_+_70517823
|
1.177
|
NM_002727
|
SRGN
|
serglycin
|
|
chr7_+_72883128
|
1.143
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr1_+_196874867
|
1.094
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr19_+_61624
|
1.046
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr12_-_101115677
|
1.044
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr3_-_173723754
|
1.043
|
NM_001190943
NM_001190942
NM_003810
|
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10
|
|
chr1_+_357502
|
1.025
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr4_+_123311207
|
1.020
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chr8_-_107023
|
1.013
|
NM_001005504
|
OR4F21
|
olfactory receptor, family 4, subfamily F, member 21
|
|
chr2_+_237143118
|
1.003
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr12_-_107551783
|
0.996
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr1_+_196874759
|
0.988
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_58899
|
0.950
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr2_+_68445936
|
0.950
|
|
PLEK
|
pleckstrin
|
|
chr1_-_148854503
|
0.891
|
|
|
|
|
chr1_-_200397300
|
0.871
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chr2_+_169631777
|
0.838
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr12_-_49708279
|
0.831
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr1_+_144152864
|
0.812
|
|
TXNIP
|
thioredoxin interacting protein
|
|
chr19_+_10689763
|
0.804
|
|
DNM2
|
dynamin 2
|
|
chr19_+_10689725
|
0.796
|
NM_001005360
NM_001005361
NM_001005362
NM_001190716
NM_004945
|
DNM2
|
dynamin 2
|
|
chr15_+_36776090
|
0.792
|
NM_207444
|
C15orf53
|
chromosome 15 open reading frame 53
|
|
chr15_-_56093477
|
0.766
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr15_-_100280784
|
0.764
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr6_-_32665409
|
0.754
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr15_+_64481270
|
0.752
|
|
MAP2K1
|
mitogen-activated protein kinase kinase 1
|
|
chr18_+_2645885
|
0.733
|
NM_015295
|
SMCHD1
|
structural maintenance of chromosomes flexible hinge domain containing 1
|
|
chr19_+_10689750
|
0.721
|
|
DNM2
|
dynamin 2
|
|
chr4_+_39084936
|
0.715
|
|
KLB
|
klotho beta
|
|
chr3_+_123279439
|
0.711
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr4_+_39084867
|
0.702
|
NM_175737
|
KLB
|
klotho beta
|
|
chr2_-_165133239
|
0.700
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr12_-_107551694
|
0.684
|
|
SELPLG
|
selectin P ligand
|
|
chr22_-_25205376
|
0.654
|
NM_152841
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr7_+_128365482
|
0.636
|
NM_032643
|
IRF5
|
interferon regulatory factor 5
|
|
chr7_+_143457083
|
0.633
|
NM_001001659
|
OR2A14
|
olfactory receptor, family 2, subfamily A, member 14
|
|
chr18_+_18989786
|
0.591
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr6_+_29382381
|
0.564
|
NM_030946
|
OR14J1
|
olfactory receptor, family 14, subfamily J, member 1
|
|
chrX_-_131181141
|
0.557
|
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr17_-_39094787
|
0.555
|
NM_001040002
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr13_+_51334117
|
0.549
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr6_-_49712483
|
0.546
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr17_+_38247493
|
0.534
|
|
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki)
|
|
chr19_+_10689813
|
0.532
|
|
DNM2
|
dynamin 2
|
|
chr11_+_63627816
|
0.532
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chr12_+_7348194
|
0.526
|
NM_001080454
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4
|
|
chr17_-_26665194
|
0.507
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_+_77939845
|
0.502
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr12_-_107551744
|
0.501
|
|
SELPLG
|
selectin P ligand
|
|
chr10_+_70517863
|
0.498
|
|
SRGN
|
serglycin
|
|
chr22_+_21393107
|
0.496
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr1_+_67404756
|
0.485
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr18_+_70317548
|
0.477
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr1_+_13903936
|
0.475
|
NM_012231
NM_015866
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr19_+_10689772
|
0.467
|
|
DNM2
|
dynamin 2
|
|
chr12_-_9159780
|
0.445
|
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_-_105169370
|
0.443
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chrX_-_77469635
|
0.438
|
NM_006639
|
CYSLTR1
|
cysteinyl leukotriene receptor 1
|
|
chr6_+_70633168
|
0.437
|
NM_001858
|
COL19A1
|
collagen, type XIX, alpha 1
|
|
chr12_-_51629874
|
0.427
|
|
KRT8
|
keratin 8
|
|
chr7_+_90177082
|
0.424
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr1_+_938709
|
0.420
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr6_+_28032997
|
0.413
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr17_+_53587492
|
0.384
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chr1_-_153477511
|
0.346
|
|
GBA
|
glucosidase, beta, acid
|
|
chr2_-_207291364
|
0.341
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr5_+_180726874
|
0.340
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr4_+_71713324
|
0.339
|
NM_031889
|
ENAM
|
enamelin
|
|
chr12_-_85756766
|
0.334
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr5_-_142795269
|
0.334
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr20_-_44575599
|
0.330
|
NM_018102
NM_199441
|
ZNF334
|
zinc finger protein 334
|
|
chr10_+_80942362
|
0.326
|
NM_001099692
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1
|
|
chr1_-_167822248
|
0.322
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr21_-_33107813
|
0.321
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr2_+_61243132
|
0.320
|
NM_001143959
|
C2orf74
|
chromosome 2 open reading frame 74
|
|
chr1_-_246525502
|
0.314
|
NM_001004692
|
OR2T12
|
olfactory receptor, family 2, subfamily T, member 12
|
|
chr2_-_74634569
|
0.314
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr2_-_135498716
|
0.306
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr2_-_2313894
|
0.306
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr1_-_154041393
|
0.301
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr5_-_22889192
|
0.300
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr20_+_5935897
|
0.291
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr4_-_83566802
|
0.288
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr7_+_44612645
|
0.288
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr6_-_25940247
|
0.286
|
NM_005074
|
SLC17A1
|
solute carrier family 17 (sodium phosphate), member 1
|
|
chr21_-_46849413
|
0.282
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr9_+_92629525
|
0.280
|
NM_001174167
|
SYK
|
spleen tyrosine kinase
|
|
chrX_-_30505804
|
0.279
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr6_+_31782618
|
0.274
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr18_-_29882555
|
0.270
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr12_-_9159742
|
0.266
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_151498802
|
0.266
|
NM_000427
|
LOR
|
loricrin
|
|
chr1_-_245237925
|
0.264
|
|
ZNF695
|
zinc finger protein 695
|
|
chr6_-_22405708
|
0.262
|
NM_000948
|
PRL
|
prolactin
|
|
chr7_-_143560850
|
0.260
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr4_-_68511738
|
0.257
|
NM_001114387
NM_182606
|
TMPRSS11A
|
transmembrane protease, serine 11A
|
|
chr2_-_43756964
|
0.257
|
NM_001101330
|
LOC728819
|
hCG1645220
|
|
chr3_-_191650291
|
0.254
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr2_-_6924092
|
0.251
|
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr3_+_112743729
|
0.248
|
|
CD96
|
CD96 molecule
|
|
chr12_-_51332214
|
0.244
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr6_-_159386007
|
0.237
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chrX_+_123062143
|
0.232
|
|
STAG2
|
stromal antigen 2
|
|
chr5_-_179040580
|
0.231
|
NM_001164444
|
CBY3
|
chibby homolog 3 (Drosophila)
|
|
chr6_-_43054865
|
0.229
|
|
PEX6
|
peroxisomal biogenesis factor 6
|
|
chr19_-_43926953
|
0.228
|
NM_144691
|
CAPN12
|
calpain 12
|
|
chr1_-_245237977
|
0.228
|
NM_020394
|
ZNF695
|
zinc finger protein 695
|
|
chr2_+_88605283
|
0.226
|
NM_152670
|
C2orf51
|
chromosome 2 open reading frame 51
|
|
chr11_-_5964790
|
0.225
|
NM_001005173
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1
|
|
chr1_-_109651140
|
0.223
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr10_+_129237602
|
0.221
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr6_+_32254108
|
0.221
|
NM_006913
|
RNF5
|
ring finger protein 5
|
|
chr3_+_82117978
|
0.220
|
|
|
|
|
chr9_-_21375258
|
0.219
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr9_+_124463665
|
0.218
|
NM_001005236
|
OR1L1
|
olfactory receptor, family 1, subfamily L, member 1
|
|
chr18_-_59137948
|
0.217
|
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr11_-_16387001
|
0.212
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_+_112743615
|
0.211
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr6_-_27988152
|
0.209
|
NM_033057
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2
|
|
chr1_-_57204208
|
0.207
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr5_-_176671897
|
0.206
|
NM_001142935
NM_031300
|
MXD3
|
MAX dimerization protein 3
|
|
chr4_-_166253453
|
0.203
|
NM_001100389
|
TMEM192
|
transmembrane protein 192
|
|
chr12_+_61645361
|
0.200
|
|
RPL14
|
ribosomal protein L14
|
|
chr11_+_71387572
|
0.197
|
NM_001039659
NM_001039660
NM_173042
|
IL18BP
|
interleukin 18 binding protein
|
|
chr5_+_140583098
|
0.195
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr11_+_73339011
|
0.188
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr1_-_167822334
|
0.184
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr18_+_70318001
|
0.184
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr20_-_62071584
|
0.182
|
NM_020713
|
ZNF512B
|
zinc finger protein 512B
|
|
chr21_+_39745649
|
0.182
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr20_-_60435983
|
0.177
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr12_+_15590811
|
0.176
|
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr10_-_46039941
|
0.175
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr6_-_166716353
|
0.171
|
|
BRP44L
|
brain protein 44-like
|
|
chr6_+_64414389
|
0.170
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr12_+_63084317
|
0.169
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr1_-_9052207
|
0.167
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr12_-_119961143
|
0.166
|
NM_003733
NM_198213
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr11_+_62813947
|
0.165
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr4_+_47403107
|
0.164
|
|
|
|
|
chr11_+_68124400
|
0.164
|
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr15_-_74091817
|
0.163
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr17_-_26665213
|
0.163
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr3_+_14691609
|
0.163
|
NM_001184957
NM_001184958
NM_032137
|
C3orf20
|
chromosome 3 open reading frame 20
|
|
chr5_+_161208475
|
0.162
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_-_153477673
|
0.162
|
NM_001171812
NM_000157
|
GBA
|
glucosidase, beta, acid
|
|
chr15_-_23204887
|
0.161
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr17_-_26665220
|
0.161
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr19_+_3713650
|
0.159
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr19_+_3713671
|
0.157
|
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr12_-_10453409
|
0.154
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 read-through transcript
killer cell lectin-like receptor subfamily C, member 4
|
|
chr9_+_102987137
|
0.154
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr21_-_44836811
|
0.154
|
NM_198688
|
KRTAP10-6
|
keratin associated protein 10-6
|
|
chr3_-_137395966
|
0.153
|
NM_001145417
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr19_-_14853166
|
0.149
|
NM_030901
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17
|
|
chr5_+_128328685
|
0.149
|
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6
|
|
chrX_-_102870207
|
0.148
|
NM_001024452
NM_001172285
|
GLRA4
|
glycine receptor, alpha 4
|
|
chr17_-_35464414
|
0.148
|
NM_001079518
NM_014815
|
MED24
|
mediator complex subunit 24
|
|
chr17_+_17548888
|
0.144
|
|
|
|
|
chr5_-_137099602
|
0.143
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr17_+_16061190
|
0.143
|
NM_004278
|
PIGL
|
phosphatidylinositol glycan anchor biosynthesis, class L
|
|
chr12_-_119826433
|
0.143
|
NM_139015
|
SPPL3
|
signal peptide peptidase 3
|
|
chr3_-_121445831
|
0.142
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr6_-_28327980
|
0.142
|
NM_019110
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4
|
|
chr7_+_63763916
|
0.141
|
NM_001013746
NM_016220
|
ZNF107
|
zinc finger protein 107
|
|
chr7_+_115650240
|
0.141
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr4_+_159351092
|
0.140
|
|
TMEM144
|
transmembrane protein 144
|
|
chr7_+_80105718
|
0.138
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr21_+_30724442
|
0.138
|
NM_181600
|
KRTAP13-4
|
keratin associated protein 13-4
|
|
chr6_-_11219932
|
0.137
|
NM_207582
|
ERVFRDE1
|
HERV-FRD provirus ancestral Env polyprotein
|
|
chr21_-_31175722
|
0.137
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr7_+_104468614
|
0.135
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr3_+_160269734
|
0.135
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr12_+_53809731
|
0.134
|
NM_001005243
|
OR9K2
|
olfactory receptor, family 9, subfamily K, member 2
|
|
chr12_+_84792203
|
0.132
|
NM_006183
|
NTS
|
neurotensin
|
|
chr6_+_26705110
|
0.132
|
NM_013375
|
ABT1
|
activator of basal transcription 1
|
|
chrX_-_110541029
|
0.131
|
NM_000555
|
DCX
|
doublecortin
|
|
chr3_+_160269836
|
0.131
|
|
IQCJ
|
IQ motif containing J
|
|
chr1_+_57093030
|
0.129
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr3_-_52544061
|
0.128
|
NM_022908
|
NT5DC2
|
5'-nucleotidase domain containing 2
|
|
chr1_-_167865973
|
0.128
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr8_-_42817604
|
0.128
|
NM_018105
NM_199003
|
THAP1
|
THAP domain containing, apoptosis associated protein 1
|
|
chr9_-_14076901
|
0.128
|
|
NFIB
|
nuclear factor I/B
|
|
chr9_-_72926333
|
0.127
|
NM_001007471
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr6_-_29163027
|
0.127
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr1_-_196776697
|
0.127
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr8_-_102034370
|
0.124
|
NM_001135699
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr1_-_245237952
|
0.123
|
|
ZNF695
|
zinc finger protein 695
|