|
chr11_+_12652427
|
4.347
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr17_-_13445942
|
3.149
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr9_-_72218098
|
2.484
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr9_+_131291144
|
2.433
|
|
|
|
|
chr9_+_131291224
|
2.351
|
|
LOC100506190
|
hypothetical LOC100506190
|
|
chr3_+_142569988
|
2.249
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr19_-_53941835
|
2.185
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr11_-_68537243
|
2.077
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr7_+_28415495
|
2.038
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr9_+_138341752
|
1.983
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr1_-_152394215
|
1.888
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chr12_+_105692466
|
1.881
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr9_+_129900590
|
1.866
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr8_+_23442331
|
1.822
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr20_+_32610161
|
1.814
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr2_-_156897286
|
1.769
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr10_+_35524835
|
1.753
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr1_-_1283770
|
1.714
|
NM_032348
|
MXRA8
|
matrix-remodelling associated 8
|
|
chr2_+_238200922
|
1.708
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr7_-_44992729
|
1.702
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr19_-_56919032
|
1.680
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr3_+_129690735
|
1.654
|
|
|
|
|
chr5_-_172130766
|
1.636
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr8_+_38363150
|
1.629
|
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr11_+_116575249
|
1.621
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr3_+_158026769
|
1.595
|
NM_001004316
NM_001193283
|
LEKR1
|
leucine, glutamate and lysine rich 1
|
|
chr10_-_62373828
|
1.575
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr2_-_216009015
|
1.499
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr17_-_7996409
|
1.485
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr19_-_7459867
|
1.476
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr6_-_127822227
|
1.469
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr9_+_133154896
|
1.452
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr6_+_136214495
|
1.428
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr6_-_53321545
|
1.418
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr4_-_186693574
|
1.412
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr4_-_140696740
|
1.410
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr8_+_23442413
|
1.404
|
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr6_+_39868116
|
1.395
|
|
|
|
|
chr1_+_27591738
|
1.385
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr3_-_188946168
|
1.382
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_+_110328850
|
1.376
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr6_+_39868126
|
1.376
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr5_+_71438796
|
1.374
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_-_53321658
|
1.365
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr14_+_95792299
|
1.364
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr3_-_101077605
|
1.348
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr12_+_10256682
|
1.331
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr16_-_78192111
|
1.325
|
NM_001031804
NM_005360
|
MAF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
|
|
chr8_+_23442307
|
1.313
|
NM_016612
|
SLC25A37
|
solute carrier family 25, member 37
|
|
chr22_+_36927884
|
1.304
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr16_+_415630
|
1.284
|
NM_014700
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr9_+_129900637
|
1.263
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr14_+_100362236
|
1.262
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr19_+_60688349
|
1.261
|
NM_020378
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative)
|
|
chr17_+_1580504
|
1.254
|
|
WDR81
|
WD repeat domain 81
|
|
chr9_+_101623957
|
1.253
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr14_+_100362204
|
1.249
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr9_-_72219359
|
1.234
|
NM_001206
|
KLF9
|
Kruppel-like factor 9
|
|
chr16_+_85101605
|
1.217
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr3_-_64186151
|
1.211
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr10_+_104525877
|
1.210
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr6_-_121697305
|
1.202
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr2_+_219852268
|
1.193
|
NM_001039550
NM_006736
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr6_-_53321706
|
1.189
|
NM_021814
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr19_-_18175731
|
1.187
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr9_+_34980266
|
1.179
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr17_-_44026042
|
1.176
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr12_+_12935619
|
1.174
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr8_+_26296439
|
1.165
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr2_+_5750229
|
1.156
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr5_+_14493905
|
1.155
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr1_+_46151845
|
1.151
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr10_+_35524060
|
1.135
|
NM_182717
NM_182718
NM_182719
NM_182720
|
CREM
|
cAMP responsive element modulator
|
|
chr11_-_118739838
|
1.128
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr8_-_145983457
|
1.116
|
NM_030580
|
ZNF34
|
zinc finger protein 34
|
|
chr16_+_415606
|
1.115
|
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr1_+_110328941
|
1.101
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr2_+_10101822
|
1.091
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr15_-_80125514
|
1.083
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr22_+_36928011
|
1.066
|
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr5_-_64813459
|
1.064
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr7_+_30140868
|
1.043
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr11_-_118740069
|
1.023
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr3_+_25444753
|
1.019
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr15_-_81037554
|
1.018
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr3_-_188945921
|
1.017
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_+_26368969
|
1.016
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr3_+_25444837
|
1.012
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr4_-_111763655
|
1.009
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr3_-_109423858
|
1.005
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr1_-_59022275
|
1.004
|
NM_002228
|
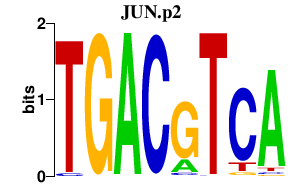
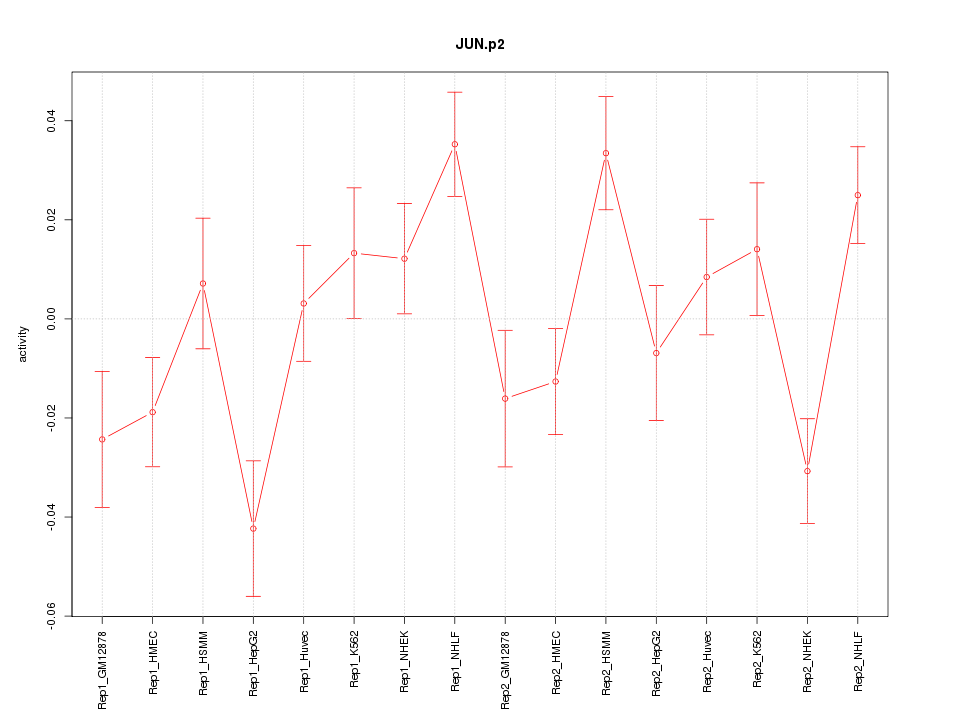
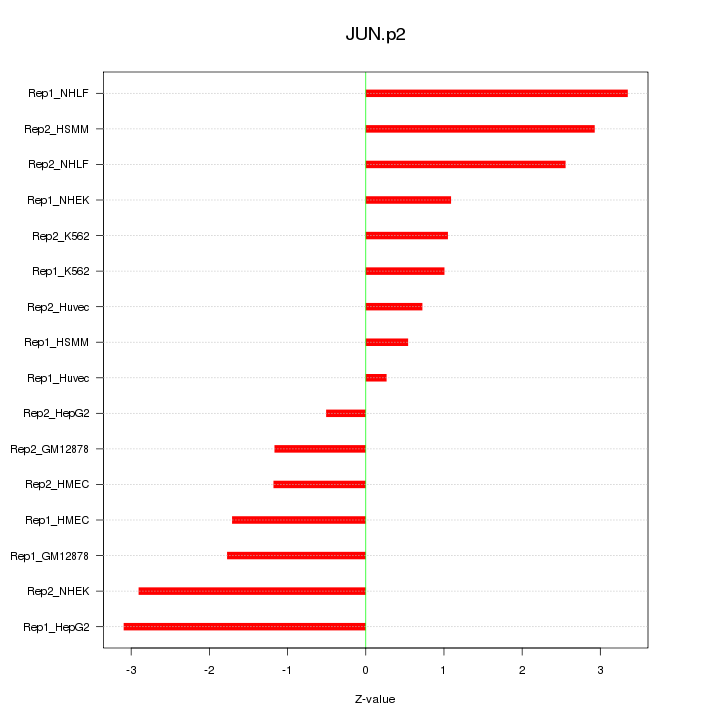
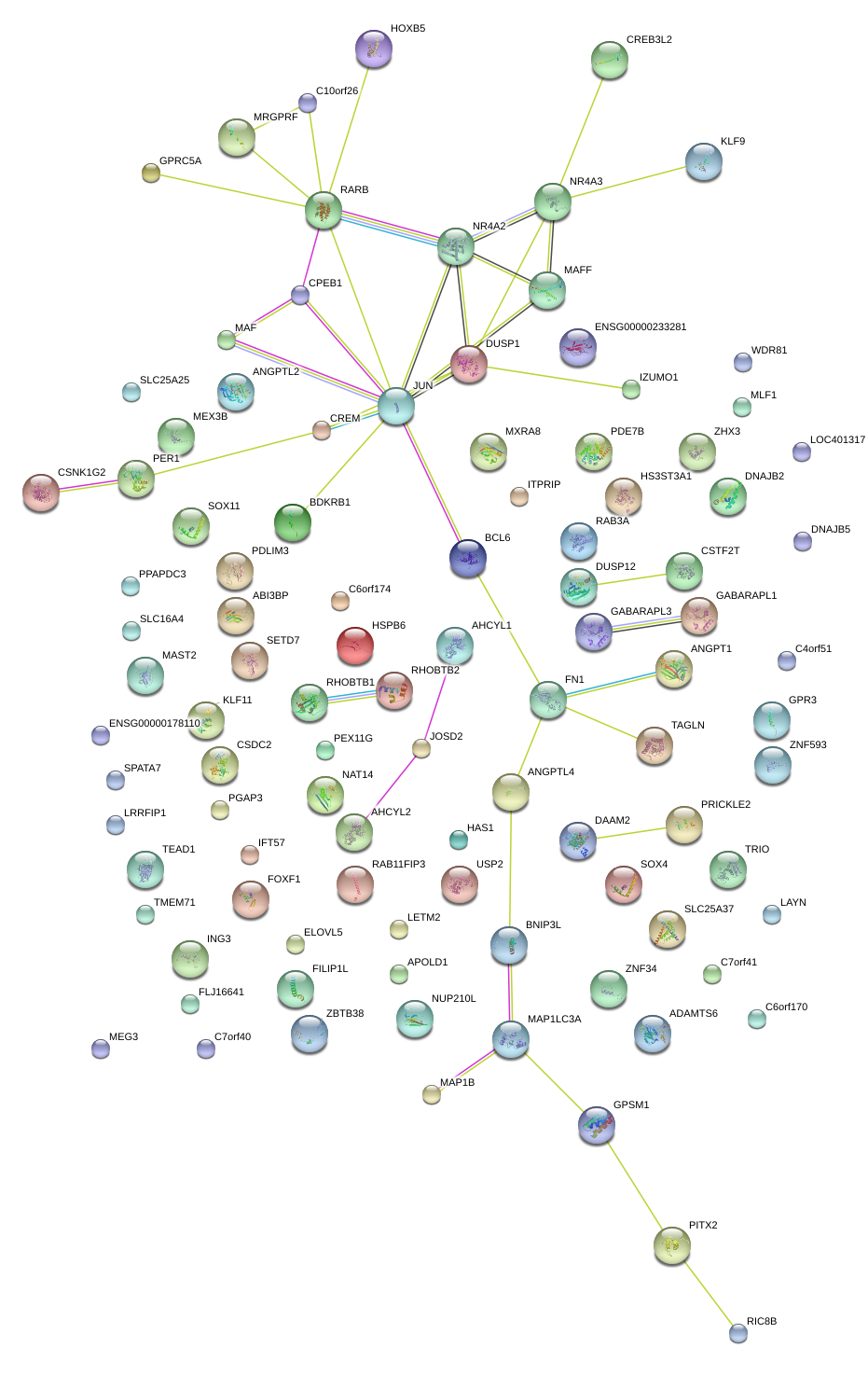
JUN
|
jun proto-oncogene
|
|
chr20_-_39379533
|
0.997
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr12_+_12829807
|
0.997
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr9_-_128924706
|
0.993
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr3_-_102194938
|
0.980
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr9_-_128924819
|
0.979
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr3_+_159771646
|
0.976
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr19_-_55706156
|
0.972
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr8_-_133841988
|
0.969
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr10_-_106083652
|
0.968
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-triphosphate receptor interacting protein
|
|
chr9_-_128924782
|
0.968
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr22_+_40286957
|
0.964
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr10_-_106088151
|
0.964
|
|
ITPRIP
|
inositol 1,4,5-triphosphate receptor interacting protein
|
|
chr12_+_12935759
|
0.957
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr14_+_87921818
|
0.950
|
|
SPATA7
|
spermatogenesis associated 7
|
|
chr4_+_146820805
|
0.946
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr7_-_137337354
|
0.944
|
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr7_-_137337352
|
0.944
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr19_-_40938251
|
0.943
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr8_-_108579270
|
0.940
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr19_+_1892184
|
0.928
|
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr10_-_53129315
|
0.925
|
NM_015235
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant
|
|
chr11_+_110916629
|
0.919
|
|
LAYN
|
layilin
|
|
chr7_+_120378052
|
0.915
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr16_+_85101734
|
0.910
|
|
FOXF1
|
forkhead box F1
|
|
chr1_+_110328955
|
0.908
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_-_110735158
|
0.902
|
NM_004696
|
SLC16A4
|
solute carrier family 16, member 4 (monocarboxylic acid transporter 5)
|
|
chr2_+_219991342
|
0.902
|
NM_001927
|
DES
|
desmin
|
|
chr1_+_110328922
|
0.901
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr7_+_22733290
|
0.898
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr11_+_119701047
|
0.897
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr2_+_30307900
|
0.896
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr12_-_113330574
|
0.890
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr4_-_111763356
|
0.888
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr14_+_75521806
|
0.882
|
NM_001102564
NM_052873
|
C14orf179
|
chromosome 14 open reading frame 179
|
|
chr14_+_87921740
|
0.880
|
NM_001040428
NM_018418
|
SPATA7
|
spermatogenesis associated 7
|
|
chr10_-_31360778
|
0.877
|
NM_001143766
NM_001143767
NM_001143768
NM_001143770
NM_001143771
NM_182755
|
ZNF438
|
zinc finger protein 438
|
|
chr10_+_102782156
|
0.875
|
|
SFXN3
|
sideroflexin 3
|
|
chr7_-_137337318
|
0.867
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chrX_+_48317772
|
0.865
|
NM_006743
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3
|
|
chr3_-_8518327
|
0.863
|
|
LOC100288428
|
hypothetical LOC100288428
|
|
chr11_+_119701214
|
0.860
|
|
TMEM136
|
transmembrane protein 136
|
|
chr12_-_62902279
|
0.859
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr4_-_111777651
|
0.858
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr6_-_91063181
|
0.857
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_+_165330710
|
0.849
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr17_+_35751796
|
0.833
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr1_+_70593073
|
0.826
|
NM_001031693
NM_001036645
NM_001036646
|
HHLA3
|
HERV-H LTR-associating 3
|
|
chr1_-_158161924
|
0.818
|
|
TAGLN2
|
transgelin 2
|
|
chr19_-_40938046
|
0.817
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr11_-_111287657
|
0.815
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr2_-_73193566
|
0.813
|
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chrX_-_83644054
|
0.810
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr5_-_54317102
|
0.806
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr13_-_35769882
|
0.804
|
|
C13orf38
|
chromosome 13 open reading frame 38
|
|
chr4_+_81406765
|
0.804
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr1_+_110328830
|
0.804
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_-_10202947
|
0.804
|
NM_004230
|
S1PR2
|
sphingosine-1-phosphate receptor 2
|
|
chr2_+_33025872
|
0.800
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr17_+_56884560
|
0.797
|
|
TBX4
|
T-box 4
|
|
chr7_-_112514990
|
0.796
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr4_-_111777584
|
0.795
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr7_+_5598979
|
0.792
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr2_+_241023703
|
0.791
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr4_+_120276386
|
0.790
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr13_-_35769968
|
0.790
|
NM_001198910
NM_001144981
NM_001144982
NM_001144983
NM_001144984
NM_001144985
NM_001144986
NM_001198908
|
C13orf38-SOHLH2
C13orf38
|
C13orf38-SOHLH2 readthrough
chromosome 13 open reading frame 38
|
|
chr19_+_1892157
|
0.785
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr7_-_112515035
|
0.783
|
|
GPR85
|
G protein-coupled receptor 85
|
|
chr3_+_25445071
|
0.782
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr3_+_8518539
|
0.777
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr18_+_75540831
|
0.773
|
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr1_-_158161902
|
0.773
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr10_+_102782151
|
0.772
|
|
SFXN3
|
sideroflexin 3
|
|
chr7_+_140042932
|
0.769
|
NM_004546
|
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa
|
|
chr7_+_100302885
|
0.768
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr21_-_42303517
|
0.768
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr6_-_29703725
|
0.766
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr15_-_80125403
|
0.766
|
NM_032246
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr21_-_38210531
|
0.760
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr20_-_33489383
|
0.758
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr4_-_123010918
|
0.755
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr9_+_35480018
|
0.748
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chrX_+_12903145
|
0.744
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr8_-_49996353
|
0.744
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr2_-_216008689
|
0.743
|
|
FN1
|
fibronectin 1
|
|
chr3_+_171558215
|
0.741
|
|
SKIL
|
SKI-like oncogene
|
|
chr7_-_112515068
|
0.739
|
NM_001146265
NM_018970
|
GPR85
|
G protein-coupled receptor 85
|
|
chr16_+_30903963
|
0.737
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr21_-_35343065
|
0.734
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_+_35479989
|
0.732
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr9_+_111927601
|
0.729
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr1_-_23758871
|
0.729
|
NM_002167
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr1_+_226394605
|
0.726
|
|
GUK1
|
guanylate kinase 1
|
|
chr14_-_88090641
|
0.726
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr3_+_189354167
|
0.725
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr18_+_75540786
|
0.725
|
NM_004715
NM_048368
|
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1
|
|
chr9_+_111582317
|
0.721
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr20_+_10147455
|
0.721
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr3_-_71715431
|
0.718
|
NM_001012505
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr5_-_131590833
|
0.711
|
NM_001017973
NM_004199
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr1_+_99502655
|
0.711
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr12_+_970613
|
0.710
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr2_-_237987509
|
0.709
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_+_226394626
|
0.709
|
|
GUK1
|
guanylate kinase 1
|
|
chr1_+_99502435
|
0.705
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr4_+_141397889
|
0.704
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr1_-_212791597
|
0.704
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr3_+_185536376
|
0.702
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr19_+_42517337
|
0.701
|
NM_181786
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member
|
|
chr22_+_17539198
|
0.699
|
|
|
|