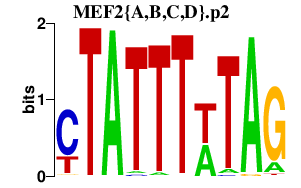
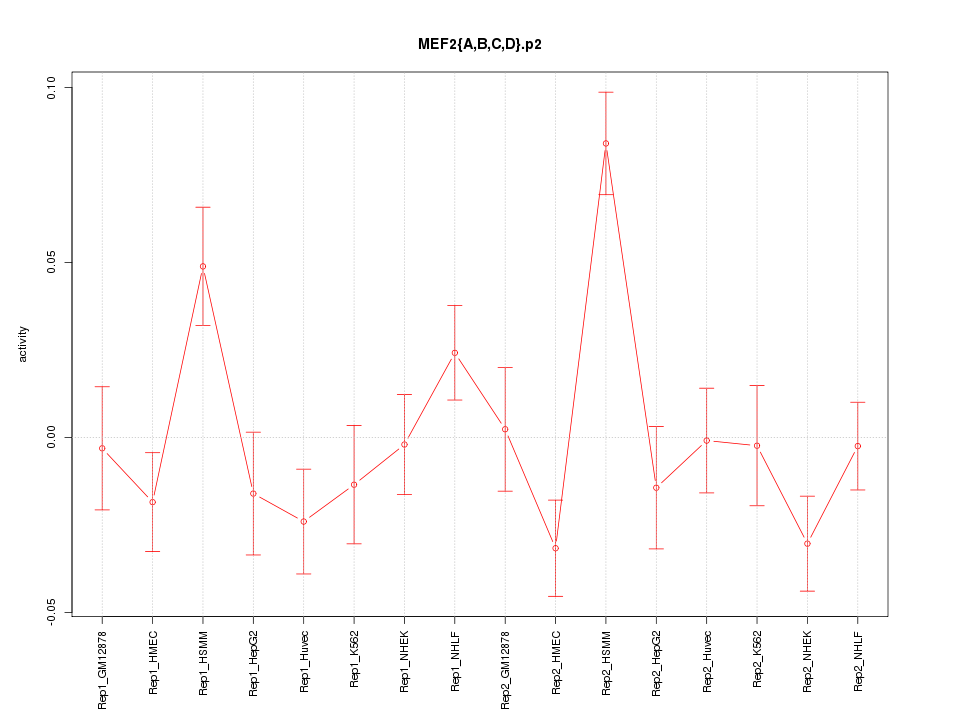
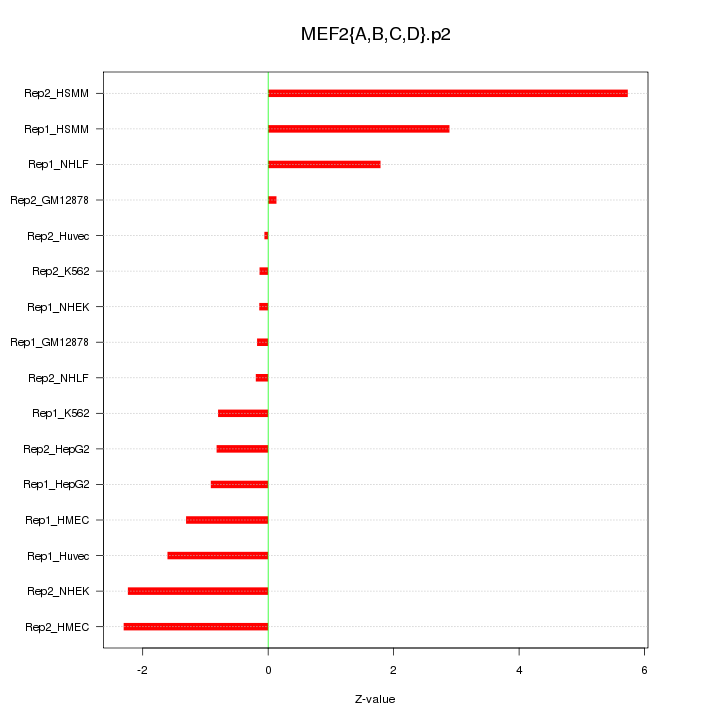
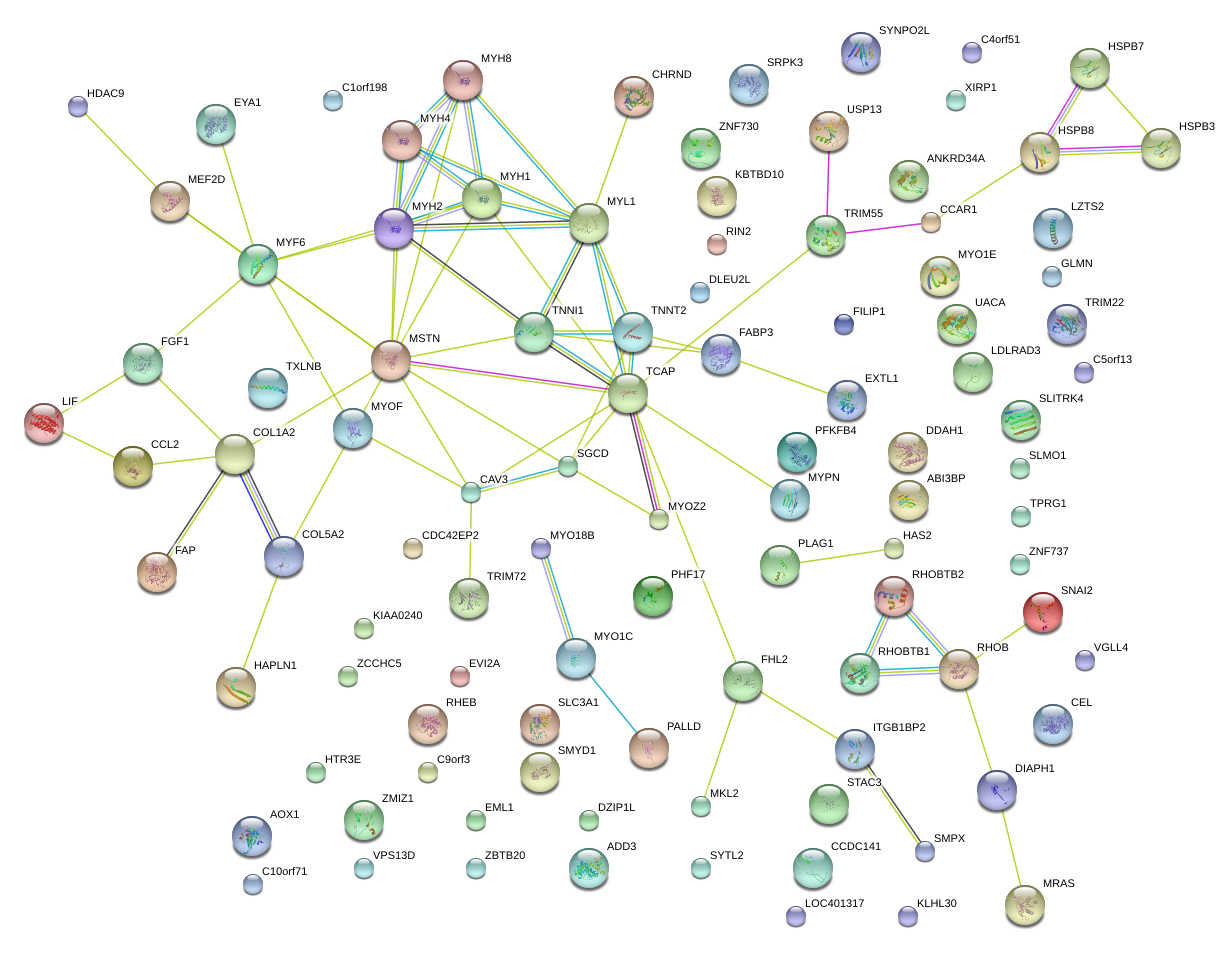
|
chr5_-_111119846
|
4.671
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_+_69539255
|
4.493
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr12_+_79625538
|
4.207
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr10_-_75080784
|
3.997
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr10_-_75085754
|
3.809
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr2_+_88148413
|
3.370
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr2_-_105421661
|
3.059
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr4_+_169654742
|
3.053
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr8_+_67201684
|
3.009
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr5_+_155686333
|
2.952
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr5_+_53787187
|
2.695
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chrX_-_77801480
|
2.623
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr16_+_31132842
|
2.596
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr6_-_139654807
|
2.539
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr2_+_170074457
|
2.498
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr15_-_68781662
|
2.300
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_-_39209075
|
2.271
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr10_+_111755715
|
2.238
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr1_-_199657468
|
2.202
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr11_-_85107569
|
2.074
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr10_+_80569464
|
2.025
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr3_-_11585264
|
1.947
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr7_+_93862137
|
1.922
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr4_+_120276386
|
1.914
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr1_-_154727014
|
1.846
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr9_+_96806235
|
1.799
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr1_-_229071957
|
1.779
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr2_+_238712101
|
1.762
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr8_-_122722674
|
1.720
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr2_-_210876560
|
1.709
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr1_-_16217025
|
1.695
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr2_-_190635566
|
1.652
|
NM_005259
|
MSTN
|
myostatin
|
|
chr8_-_49996353
|
1.644
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr20_+_19903778
|
1.628
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr12_+_118100977
|
1.619
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr7_+_28415495
|
1.596
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr16_+_14187965
|
1.595
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr3_-_115825742
|
1.561
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr22_+_24468168
|
1.532
|
|
MYO18B
|
myosin XVIIIB
|
|
chrX_+_152699616
|
1.529
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr2_-_162808123
|
1.519
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr17_+_35075124
|
1.501
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chrX_-_21686079
|
1.491
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chrX_-_142549984
|
1.477
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr4_+_146820805
|
1.427
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr1_-_85816520
|
1.420
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr10_-_62373828
|
1.404
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr19_+_23091616
|
1.400
|
|
ZNF730
|
zinc finger protein 730
|
|
chr22_+_24468110
|
1.379
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr18_+_12397894
|
1.357
|
NM_001142405
NM_006553
|
SLMO1
|
slowmo homolog 1 (Drosophila)
|
|
chr1_-_199613427
|
1.354
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr1_-_31618405
|
1.353
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr1_-_199613406
|
1.347
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr12_-_55931172
|
1.345
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr7_+_18501876
|
1.319
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_102194938
|
1.317
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr2_-_179623030
|
1.303
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr14_+_99309688
|
1.296
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr3_+_139550368
|
1.291
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr10_+_50177192
|
1.280
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr11_+_35922106
|
1.273
|
NM_174902
|
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr2_-_189752575
|
1.258
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr22_-_28972683
|
1.253
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr5_-_83052606
|
1.252
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr3_+_8750485
|
1.212
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr11_+_5667494
|
1.167
|
NM_006074
|
TRIM22
|
tripartite motif containing 22
|
|
chr1_+_144183811
|
1.149
|
|
ANKRD34A
|
ankyrin repeat domain 34A
|
|
chr1_+_12325632
|
1.137
|
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae)
|
|
chr3_-_48573556
|
1.132
|
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr3_+_139550134
|
1.129
|
NM_001085049
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr1_+_26220852
|
1.110
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr4_-_1185144
|
1.082
|
|
LOC100130872
|
hypothetical LOC100130872
|
|
chr6_+_42896771
|
1.068
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr2_+_233099113
|
1.027
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr11_+_64838882
|
1.011
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr8_-_72431274
|
1.004
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_95231933
|
0.986
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr11_+_5667392
|
0.984
|
|
TRIM22
|
tripartite motif containing 22
|
|
chr10_+_102748062
|
0.980
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr19_-_20540380
|
0.973
|
NM_001159293
|
ZNF737
|
zinc finger protein 737
|
|
chr3_+_190147633
|
0.972
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr3_+_180853473
|
0.971
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr2_+_44356100
|
0.964
|
NM_000341
|
SLC3A1
|
solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1
|
|
chr2_+_20510312
|
0.951
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr17_-_26672757
|
0.942
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr6_-_76259976
|
0.939
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr17_-_1335763
|
0.933
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr17_+_29606408
|
0.916
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr19_+_43847049
|
0.916
|
|
|
|
|
chr4_+_129950222
|
0.914
|
NM_024900
NM_199320
|
PHF17
|
PHD finger protein 17
|
|
chr4_+_169789327
|
0.904
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr10_+_70172257
|
0.901
|
|
CCAR1
|
cell division cycle and apoptosis regulator 1
|
|
chr2_+_201182187
|
0.873
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr3_+_185300660
|
0.854
|
NM_182589
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3, family member E
|
|
chr2_+_37282516
|
0.854
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chrX_+_70438352
|
0.851
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr3_-_139317140
|
0.847
|
NM_173543
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr5_-_140875591
|
0.833
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr13_-_49597776
|
0.828
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr5_-_142057692
|
0.821
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr17_-_10265991
|
0.805
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr13_-_59223074
|
0.793
|
|
|
|
|
chr1_+_26220895
|
0.791
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr7_-_150847884
|
0.786
|
NM_005614
|
RHEB
|
Ras homolog enriched in brain
|
|
chr17_+_65677255
|
0.785
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr15_+_61676604
|
0.777
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr3_-_113842652
|
0.774
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr14_+_23670514
|
0.769
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr2_+_149603226
|
0.767
|
NM_177964
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr14_+_95792299
|
0.761
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr9_+_131123115
|
0.754
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr9_+_70161634
|
0.754
|
NM_021965
|
PGM5
|
phosphoglucomutase 5
|
|
chr14_-_88090641
|
0.753
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr11_+_1897374
|
0.753
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr5_+_244625
|
0.748
|
NM_001080478
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr9_+_115396020
|
0.746
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr8_+_75066117
|
0.745
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr10_-_92671005
|
0.732
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr17_-_24491521
|
0.731
|
|
MYO18A
|
myosin XVIIIA
|
|
chr2_+_220007943
|
0.729
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr6_-_130224108
|
0.727
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr5_+_132037271
|
0.720
|
NM_000589
NM_172348
|
IL4
|
interleukin 4
|
|
chr9_-_101621964
|
0.720
|
|
|
|
|
chr2_+_85774924
|
0.714
|
NM_006433
NM_012483
|
GNLY
|
granulysin
|
|
chr6_+_136214495
|
0.708
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr4_-_177427252
|
0.706
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr3_+_100840129
|
0.703
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr17_-_1336073
|
0.702
|
|
MYO1C
|
myosin IC
|
|
chr4_-_186934059
|
0.699
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr16_-_73830480
|
0.695
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr21_+_29439768
|
0.691
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr1_+_168899936
|
0.689
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr21_-_35174742
|
0.681
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr15_+_76171981
|
0.680
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr4_-_138673078
|
0.679
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chrX_+_109548940
|
0.676
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr7_+_18502342
|
0.669
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr9_-_21218132
|
0.662
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr15_+_66695932
|
0.660
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr16_-_31347151
|
0.660
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr10_+_91082218
|
0.659
|
NM_001031683
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr2_+_5750229
|
0.649
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr17_-_77474597
|
0.647
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr9_+_126094388
|
0.642
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr12_+_119224506
|
0.633
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr1_+_36394044
|
0.624
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr10_-_69095362
|
0.621
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr15_-_70838703
|
0.616
|
|
|
|
|
chr17_-_45626364
|
0.612
|
|
|
|
|
chr3_+_8518539
|
0.607
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr7_+_30917939
|
0.604
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr3_+_35696119
|
0.597
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr2_+_69055208
|
0.596
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr8_+_1980564
|
0.594
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr19_-_53305825
|
0.587
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr9_+_126094067
|
0.584
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr5_-_150501393
|
0.578
|
NM_001193544
|
ANXA6
|
annexin A6
|
|
chr17_+_39603599
|
0.574
|
NM_080863
|
ASB16
|
ankyrin repeat and SOCS box containing 16
|
|
chr2_-_207738858
|
0.574
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr4_-_157094443
|
0.574
|
NM_001334
|
CTSO
|
cathepsin O
|
|
chr15_+_40427592
|
0.573
|
NM_212465
|
CAPN3
|
calpain 3, (p94)
|
|
chr4_-_5881271
|
0.570
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr7_+_63892198
|
0.568
|
NM_001160183
NM_006524
|
ZNF138
|
zinc finger protein 138
|
|
chr3_+_42702014
|
0.565
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr10_-_69125932
|
0.558
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr19_+_20750949
|
0.557
|
|
|
|
|
chr3_-_55489973
|
0.551
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr3_+_110024234
|
0.550
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr1_+_154478574
|
0.549
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr5_-_149515502
|
0.547
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr9_+_130257254
|
0.541
|
NM_153436
|
ODF2
|
outer dense fiber of sperm tails 2
|
|
chr3_-_127810087
|
0.538
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr10_+_32896656
|
0.535
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr7_-_76667045
|
0.534
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr11_-_62213577
|
0.533
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr10_+_86174665
|
0.532
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr12_-_242255
|
0.531
|
NM_001190997
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr1_+_36394338
|
0.531
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr10_+_91142301
|
0.521
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr4_-_170428681
|
0.517
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr13_-_20533703
|
0.515
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_-_8485026
|
0.514
|
NM_001146175
NM_032370
|
ZNF414
|
zinc finger protein 414
|
|
chr19_+_19872700
|
0.514
|
NM_031218
|
ZNF93
|
zinc finger protein 93
|
|
chr1_-_151784889
|
0.510
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr5_-_149515331
|
0.508
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_21985433
|
0.508
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr1_-_115039614
|
0.507
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr3_-_113842553
|
0.507
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr2_+_219991342
|
0.505
|
NM_001927
|
DES
|
desmin
|
|
chr11_+_5597749
|
0.504
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr16_-_66535457
|
0.499
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr1_+_144236346
|
0.497
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr9_+_81377289
|
0.493
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr9_-_74169649
|
0.492
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr17_-_43254117
|
0.491
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr10_+_88418243
|
0.477
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr1_+_144236398
|
0.473
|
|
ITGA10
|
integrin, alpha 10
|
|
chr3_+_44641505
|
0.471
|
NM_001024855
NM_006991
|
ZNF197
|
zinc finger protein 197
|
|
chr12_-_120781002
|
0.469
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr9_-_14769530
|
0.467
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|