|
chr6_+_45498354
|
4.515
|
|
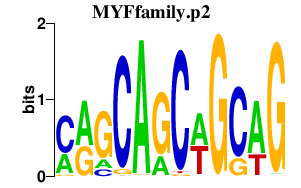
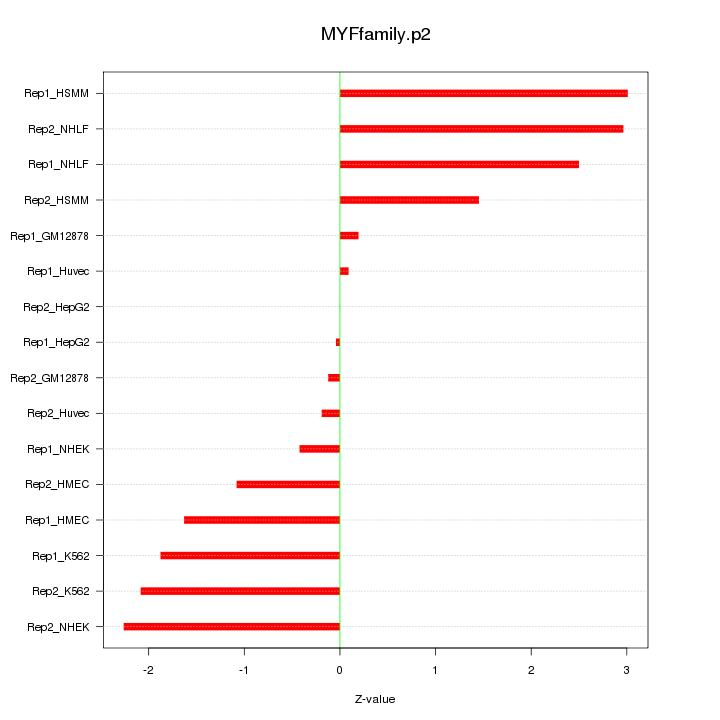
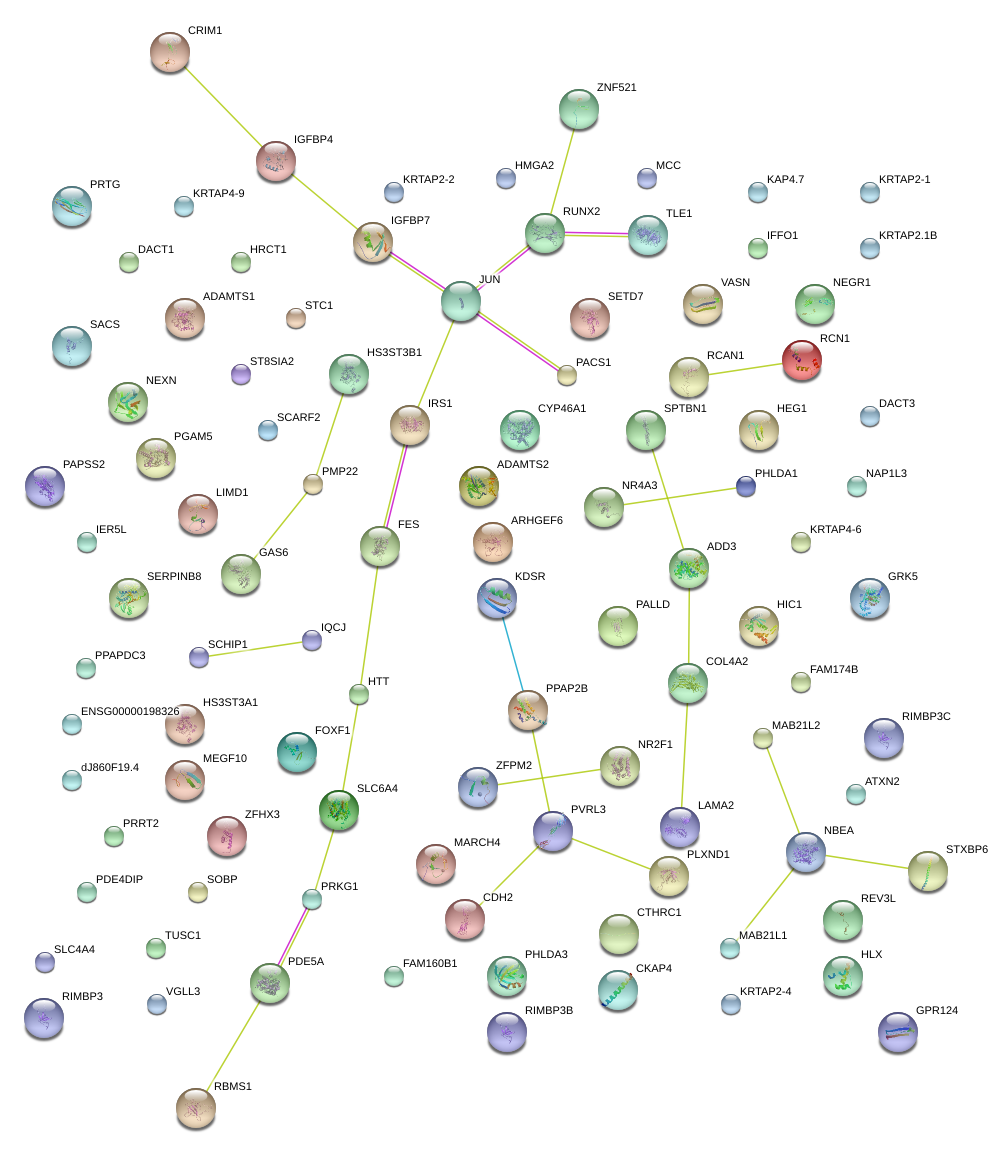
RUNX2
|
runt-related transcription factor 2
|
|
chr6_+_45498262
|
3.396
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr12_-_74710741
|
2.400
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr6_+_129245978
|
1.992
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr6_+_107917965
|
1.825
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr4_+_3046034
|
1.775
|
NM_002111
|
HTT
|
huntingtin
|
|
chr18_+_59788242
|
1.714
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr2_-_227372574
|
1.707
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_+_35896188
|
1.670
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr14_+_58174489
|
1.610
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr1_-_72521079
|
1.588
|
|
|
|
|
chr5_-_178705036
|
1.503
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr3_-_126257425
|
1.430
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr17_+_14145051
|
1.412
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr1_-_56817802
|
1.402
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_-_74711474
|
1.392
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr13_-_34948758
|
1.378
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr22_+_20068039
|
1.374
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr10_+_89409332
|
1.338
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr18_-_21185968
|
1.329
|
|
ZNF521
|
zinc finger protein 521
|
|
chrX_-_92815222
|
1.313
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr1_+_78126787
|
1.309
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr17_-_15104768
|
1.300
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr9_-_130980302
|
1.279
|
|
IER5L
|
immediate early response 5-like
|
|
chr22_-_20235372
|
1.265
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr5_-_178704934
|
1.251
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr12_-_105165805
|
1.218
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr17_+_1906262
|
1.206
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr12_-_74711642
|
1.175
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr10_+_120957084
|
1.169
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr14_-_24588916
|
1.166
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr17_+_36493927
|
1.163
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr18_-_24010944
|
1.138
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr2_-_227371698
|
1.132
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr16_-_71650034
|
1.126
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr13_+_34948885
|
1.111
|
|
NBEA
|
neurobeachin
|
|
chr22_-_19122047
|
1.096
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr8_-_23768242
|
1.081
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr18_-_21186107
|
1.079
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr3_+_160964574
|
1.058
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_64504835
|
1.052
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr1_-_72520735
|
1.048
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr4_-_140696740
|
1.046
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chrX_-_135677022
|
1.045
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr8_+_106400322
|
1.037
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr2_-_161058120
|
1.021
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr16_+_85101734
|
1.004
|
|
FOXF1
|
forkhead box F1
|
|
chr17_+_36515166
|
0.983
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr1_+_219119623
|
0.973
|
|
HLX
|
H2.0-like homeobox
|
|
chr4_-_57671273
|
0.969
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_57671112
|
0.967
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr3_-_130807865
|
0.961
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr4_-_57671307
|
0.953
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr1_-_56817568
|
0.946
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_-_56817717
|
0.944
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_29731058
|
0.942
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr8_+_104452961
|
0.940
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr15_-_53822468
|
0.939
|
NM_173814
|
PRTG
|
protogenin
|
|
chr16_+_85101605
|
0.939
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr10_+_111755715
|
0.938
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr20_+_2743613
|
0.923
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr4_+_159311353
|
0.918
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr3_+_112273352
|
0.917
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr17_-_36469869
|
0.910
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr2_+_54536780
|
0.907
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr6_-_111911029
|
0.893
|
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chr3_+_112273464
|
0.883
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr12_-_6535481
|
0.880
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr12_-_110521407
|
0.870
|
|
ATXN2
|
ataxin 2
|
|
chr3_+_45611180
|
0.860
|
NM_014240
|
LIMD1
|
LIM domains containing 1
|
|
chr2_-_216944911
|
0.851
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr13_+_109757593
|
0.849
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr2_+_36436873
|
0.843
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr11_+_65594361
|
0.839
|
NM_018026
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr15_-_90999606
|
0.839
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr9_+_101623957
|
0.833
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_-_199704773
|
0.825
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr10_+_116571485
|
0.824
|
NM_001135051
NM_020940
|
FAM160B1
|
family with sequence similarity 160, member B1
|
|
chr9_-_83494198
|
0.821
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr17_+_35853177
|
0.814
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr3_-_87122936
|
0.808
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr2_-_161058550
|
0.808
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr8_+_37773991
|
0.804
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr15_+_89229275
|
0.802
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr15_+_90738108
|
0.800
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr8_+_104452877
|
0.798
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr14_+_99220349
|
0.791
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr17_-_36475652
|
0.786
|
NM_033184
|
KRTAP2-4
|
keratin associated protein 2-4
|
|
chr5_+_92946348
|
0.785
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr12_+_62523648
|
0.781
|
|
|
|
|
chr4_-_120769275
|
0.768
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr13_+_113546912
|
0.761
|
|
GAS6
|
growth arrest-specific 6
|
|
chr5_-_112852425
|
0.757
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr1_-_59020875
|
0.754
|
|
JUN
|
jun proto-oncogene
|
|
chr21_-_27139143
|
0.751
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr9_-_130980359
|
0.748
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr13_-_22905822
|
0.747
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr16_+_4370879
|
0.745
|
|
VASN
|
vasorin
|
|
chr11_+_32069183
|
0.744
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr4_+_72271218
|
0.737
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr4_+_169989680
|
0.735
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr5_+_126654354
|
0.734
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr9_+_133154896
|
0.728
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr18_-_59185330
|
0.728
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr9_-_25668230
|
0.726
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr3_-_87122958
|
0.719
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr10_+_52504239
|
0.716
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr1_-_143786993
|
0.716
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_+_26239755
|
0.710
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr2_+_187266899
|
0.708
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr21_+_46226072
|
0.708
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr2_-_217267742
|
0.707
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr12_+_6807831
|
0.707
|
NM_014262
|
LEPREL2
|
leprecan-like 2
|
|
chr6_-_91063181
|
0.704
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_-_160972606
|
0.702
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr15_+_89228668
|
0.702
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr9_-_128924706
|
0.700
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr17_-_36456949
|
0.700
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr10_+_90629880
|
0.699
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr4_-_142273881
|
0.696
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr8_-_72918697
|
0.695
|
|
MSC
|
musculin
|
|
chr9_-_128924782
|
0.693
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr9_+_77695334
|
0.693
|
NM_001190482
NM_006200
|
PCSK5
|
proprotein convertase subtilisin/kexin type 5
|
|
chr9_+_132700567
|
0.688
|
NM_005157
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr6_-_56816403
|
0.687
|
|
DST
|
dystonin
|
|
chr15_-_46724360
|
0.687
|
|
FBN1
|
fibrillin 1
|
|
chr7_+_93861808
|
0.685
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr3_-_73756668
|
0.685
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr19_-_50977592
|
0.673
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr6_-_31347867
|
0.672
|
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chrX_+_53095230
|
0.667
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr6_-_31347805
|
0.664
|
NM_002117
|
HLA-C
|
major histocompatibility complex, class I, C
|
|
chrX_-_19815526
|
0.664
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_-_120769590
|
0.660
|
|
|
|
|
chr2_+_191453791
|
0.659
|
NM_014905
|
GLS
|
glutaminase
|
|
chr13_-_109236897
|
0.659
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr19_-_50977586
|
0.658
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_+_132700462
|
0.653
|
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr14_-_26136799
|
0.650
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr17_+_58058774
|
0.642
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr22_+_19699441
|
0.640
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr9_+_136673534
|
0.639
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr1_+_216586199
|
0.635
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chrX_+_46318135
|
0.634
|
NM_019886
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7
|
|
chr8_+_37672427
|
0.632
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr2_+_191453813
|
0.632
|
|
GLS
|
glutaminase
|
|
chr19_-_50977485
|
0.630
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr7_+_90731671
|
0.628
|
NM_003505
|
FZD1
|
frizzled homolog 1 (Drosophila)
|
|
chr9_-_128924819
|
0.627
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr16_-_73198416
|
0.624
|
|
GLG1
|
golgi glycoprotein 1
|
|
chr10_+_124210980
|
0.623
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr6_-_111911106
|
0.619
|
NM_002912
|
REV3L
|
REV3-like, catalytic subunit of DNA polymerase zeta (yeast)
|
|
chrX_+_109548940
|
0.619
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr16_-_30983909
|
0.617
|
NM_001172669
NM_001172670
|
ZNF668
|
zinc finger protein 668
|
|
chr14_+_101500650
|
0.613
|
|
DYNC1H1
|
dynein, cytoplasmic 1, heavy chain 1
|
|
chr3_-_49370682
|
0.613
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr12_-_54387934
|
0.612
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr13_+_113546943
|
0.611
|
|
GAS6
|
growth arrest-specific 6
|
|
chr1_+_67163165
|
0.609
|
NM_001077700
NM_001077702
NM_001077703
NM_001146110
NM_001146111
NM_001146112
NM_001146113
NM_020948
|
MIER1
|
mesoderm induction early response 1 homolog (Xenopus laevis)
|
|
chr17_+_72883759
|
0.603
|
NM_001113494
|
SEPT9
|
septin 9
|
|
chr13_+_30378311
|
0.596
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr1_-_36589744
|
0.595
|
|
|
|
|
chr6_+_114285209
|
0.595
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr1_-_29321599
|
0.592
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr9_+_98252200
|
0.582
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr22_+_44828338
|
0.580
|
|
|
|
|
chr16_-_73830480
|
0.577
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chrX_-_19815370
|
0.576
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr4_+_14614462
|
0.573
|
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr4_+_2783764
|
0.570
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr14_-_74148422
|
0.567
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr11_-_6298284
|
0.567
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr12_+_26239535
|
0.566
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr4_+_109072120
|
0.565
|
NM_183075
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1
|
|
chr7_+_93862137
|
0.565
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr11_-_10272256
|
0.555
|
NM_030962
|
SBF2
|
SET binding factor 2
|
|
chr3_-_147361465
|
0.553
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr20_-_33489383
|
0.548
|
NM_000557
|
GDF5
|
growth differentiation factor 5
|
|
chr1_-_32479785
|
0.540
|
|
MTMR9LP
|
myotubularin related protein 9-like, pseudogene
|
|
chr1_-_35431318
|
0.539
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr18_+_19848749
|
0.539
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr5_-_59225377
|
0.538
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_54387732
|
0.538
|
|
ITGA7
|
integrin, alpha 7
|
|
chr1_+_167342149
|
0.537
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr18_-_53621277
|
0.537
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr6_-_143307976
|
0.535
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr20_+_43953428
|
0.533
|
|
CTSA
|
cathepsin A
|
|
chr3_-_49424506
|
0.529
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr22_+_19248733
|
0.528
|
|
MED15
|
mediator complex subunit 15
|
|
chr8_-_72918938
|
0.527
|
|
MSC
|
musculin
|
|
chr12_-_88270128
|
0.523
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr18_-_24011349
|
0.522
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr13_+_113546897
|
0.522
|
|
GAS6
|
growth arrest-specific 6
|
|
chr17_-_74433048
|
0.521
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr11_-_72698050
|
0.520
|
|
LOC100287837
|
hypothetical protein LOC100287837
|
|
chr4_-_17632456
|
0.520
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr10_+_71833866
|
0.520
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr4_+_81325447
|
0.519
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr11_+_117982422
|
0.518
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr10_+_71833905
|
0.514
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|