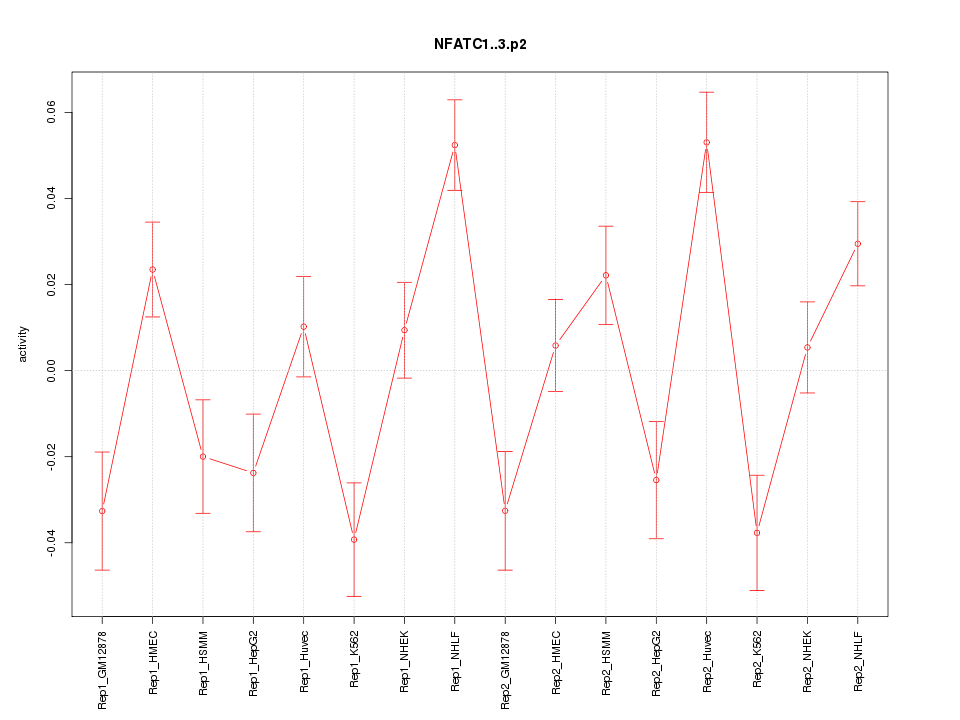
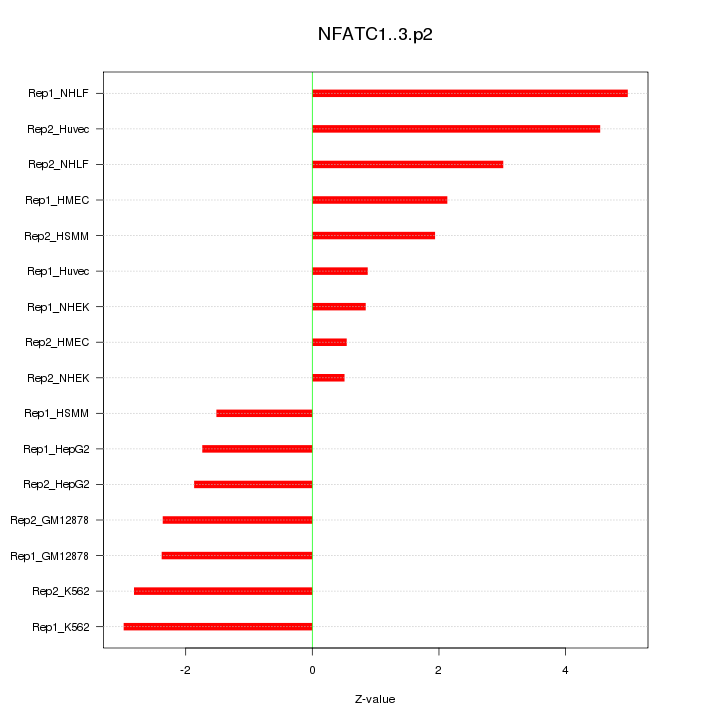
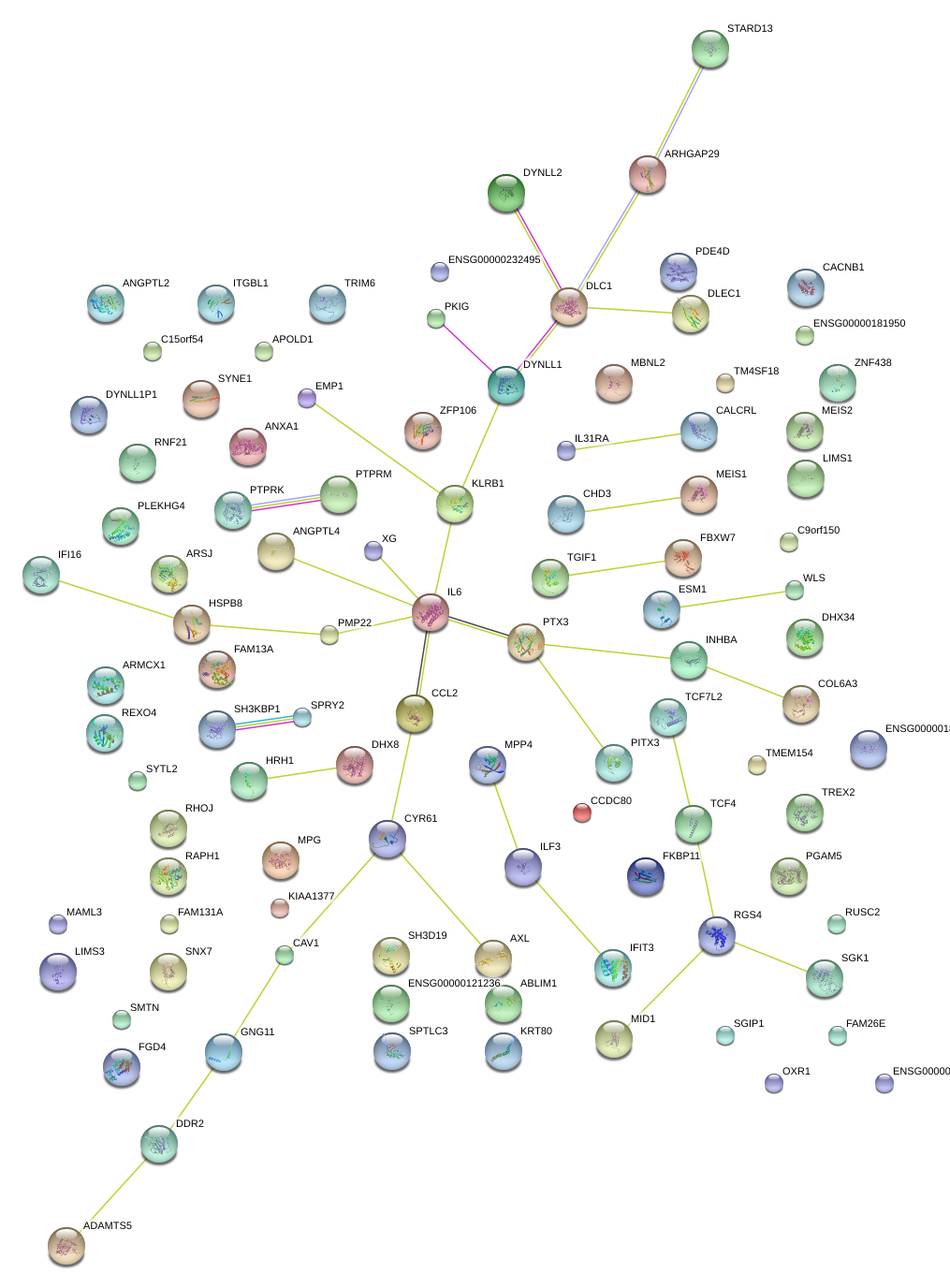
|
chr12_+_13240983
|
9.825
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240987
|
9.559
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240868
|
8.217
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr10_-_116434364
|
6.670
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr18_-_51219880
|
5.569
|
|
TCF4
|
transcription factor 4
|
|
chr10_-_116434102
|
5.049
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_-_85115105
|
4.469
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr8_-_13416554
|
4.385
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_-_128883125
|
4.054
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr22_-_34566210
|
3.892
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_157246305
|
3.871
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr9_+_676640
|
3.694
|
|
|
|
|
chr1_+_160868932
|
3.454
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr15_-_40537002
|
3.363
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr4_-_115120251
|
3.329
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr13_-_79810972
|
3.301
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr22_-_34566367
|
3.200
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr22_-_34566276
|
3.186
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_160868822
|
3.114
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr18_+_3443771
|
2.963
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr4_-_152366970
|
2.805
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_-_50871953
|
2.803
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr11_+_65027721
|
2.791
|
|
|
|
|
chr4_-_153820535
|
2.729
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr12_+_118100977
|
2.726
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr14_+_62740878
|
2.701
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr3_+_185537969
|
2.677
|
NM_144635
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr2_-_202271598
|
2.670
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr2_-_188021117
|
2.658
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr9_+_35528890
|
2.600
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr9_+_74956494
|
2.588
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr4_-_141293572
|
2.544
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr12_+_32578513
|
2.507
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chrX_-_19598912
|
2.474
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_66772393
|
2.437
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chrX_-_10504852
|
2.425
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chrX_+_100692071
|
2.422
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr17_+_7732759
|
2.409
|
NM_001005273
NM_005852
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr20_+_12937616
|
2.392
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr11_+_101290930
|
2.388
|
NM_020802
|
KIAA1377
|
KIAA1377
|
|
chr7_+_115952074
|
2.361
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr6_+_116939500
|
2.349
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr6_-_152744022
|
2.309
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr15_+_37330176
|
2.299
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chr9_+_12764988
|
2.275
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr2_+_66519949
|
2.266
|
|
MEIS1
|
Meis homeobox 1
|
|
chr3_-_150533948
|
2.236
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr9_+_35528628
|
2.222
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr1_-_68470585
|
2.211
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr12_+_12829807
|
2.200
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr10_-_31328426
|
2.183
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr4_-_152368492
|
2.176
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr7_+_93388951
|
2.146
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr20_+_42644633
|
2.142
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr3_-_194118321
|
2.121
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr4_-_153493133
|
2.107
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr13_+_100902942
|
2.100
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr5_-_54317102
|
2.094
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chrX_+_2680092
|
2.059
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chr3_-_113842553
|
2.040
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_22733626
|
2.015
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr1_+_85819004
|
2.009
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr17_+_29606408
|
2.003
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr21_-_27261276
|
2.002
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr3_-_113842652
|
1.991
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_+_56772391
|
1.974
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr13_+_96672541
|
1.938
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr13_-_32822759
|
1.931
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_158637273
|
1.920
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr5_+_55182963
|
1.909
|
NM_139017
|
IL31RA
|
interleukin 31 receptor A
|
|
chr7_-_41709191
|
1.903
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr3_+_11242666
|
1.871
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr1_-_94475504
|
1.867
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_+_98899734
|
1.860
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr11_+_5597749
|
1.845
|
NM_001003827
|
TRIM34
|
tripartite motif containing 34
|
|
chr2_-_237987545
|
1.844
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_-_15104768
|
1.842
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr2_-_237987553
|
1.833
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr9_-_128924819
|
1.824
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr18_+_7936885
|
1.823
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr1_+_161305558
|
1.821
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_+_29808845
|
1.800
|
|
SMTN
|
smoothelin
|
|
chr2_+_110013297
|
1.772
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr8_+_107351519
|
1.770
|
NM_001198533
NM_018002
|
LOC346887
OXR1
|
similar to solute carrier family 16 (monocarboxylic acid transporters), member 14
oxidation resistance 1
|
|
chr10_+_91077581
|
1.768
|
NM_001549
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3
|
|
chr19_+_46417117
|
1.768
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr6_-_134537699
|
1.750
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr6_-_134537637
|
1.743
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr4_-_89963177
|
1.723
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr5_-_59100091
|
1.723
|
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_204108151
|
1.722
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr2_-_110665188
|
1.708
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr16_+_65868913
|
1.699
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr11_+_113672407
|
1.683
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr18_+_64616296
|
1.678
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr12_+_10074542
|
1.657
|
NM_207345
|
CLEC9A
|
C-type lectin domain family 9, member A
|
|
chr7_+_28692122
|
1.652
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr11_-_66898223
|
1.651
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr5_-_59100194
|
1.638
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_+_34720368
|
1.615
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr2_-_237987509
|
1.602
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr6_-_2580296
|
1.600
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr9_+_5500544
|
1.585
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr20_+_19863716
|
1.583
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_-_158985727
|
1.571
|
NM_006519
|
DYNLT1
|
dynein, light chain, Tctex-type 1
|
|
chr3_+_103029523
|
1.571
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr13_-_22905822
|
1.558
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr2_-_237987579
|
1.556
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr8_+_42671718
|
1.556
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr2_-_237987554
|
1.552
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_94475840
|
1.551
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_-_151804929
|
1.549
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr11_-_68537243
|
1.544
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr20_-_14266200
|
1.542
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr3_-_113841448
|
1.539
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr17_-_44022595
|
1.539
|
|
HOXB3
|
homeobox B3
|
|
chr16_+_4842077
|
1.517
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr5_-_73973004
|
1.507
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr5_-_146813343
|
1.495
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr6_-_112682401
|
1.491
|
|
LAMA4
|
laminin, alpha 4
|
|
chr8_-_93144039
|
1.490
|
|
|
|
|
chr17_-_30783631
|
1.489
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr2_+_108637698
|
1.488
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr2_-_189752575
|
1.487
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr4_-_70661018
|
1.484
|
NM_014465
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1
|
|
chr3_+_69216747
|
1.470
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr1_-_79244948
|
1.464
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr18_+_27281729
|
1.464
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr18_+_3441589
|
1.462
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr6_-_112186977
|
1.459
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr1_-_89261021
|
1.458
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr17_-_40339919
|
1.455
|
|
GFAP
|
glial fibrillary acidic protein
|
|
chr8_-_49996353
|
1.443
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr11_-_71458739
|
1.436
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr12_-_90097384
|
1.431
|
NM_133503
|
DCN
|
decorin
|
|
chr2_-_227952249
|
1.416
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr6_+_116798791
|
1.416
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr19_+_42689881
|
1.412
|
|
ZNF793
|
zinc finger protein 793
|
|
chr6_-_33862690
|
1.411
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr2_-_55090759
|
1.407
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr11_-_104344473
|
1.406
|
NM_001225
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chr3_-_124893875
|
1.406
|
|
MYLK
|
myosin light chain kinase
|
|
chr4_+_109033868
|
1.406
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_-_109423858
|
1.406
|
NM_018010
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas)
|
|
chr17_-_5428491
|
1.400
|
NM_001033053
NM_014922
NM_033004
NM_033006
NM_033007
|
NLRP1
|
NLR family, pyrin domain containing 1
|
|
chr16_-_63713485
|
1.386
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_+_46151845
|
1.383
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr21_-_38792173
|
1.381
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr2_-_201930311
|
1.380
|
NM_001127391
NM_139163
|
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12
|
|
chr16_-_63713382
|
1.374
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr16_-_63713466
|
1.373
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_+_215871288
|
1.369
|
NM_138796
|
SPATA17
|
spermatogenesis associated 17
|
|
chr6_-_112682459
|
1.368
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr21_-_35174742
|
1.368
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr5_-_58607673
|
1.360
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr20_-_17487480
|
1.354
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr9_+_88953378
|
1.349
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr5_+_38481392
|
1.344
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr12_-_48902692
|
1.339
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr1_-_215870935
|
1.336
|
NM_018040
|
GPATCH2
|
G patch domain containing 2
|
|
chr11_-_35503720
|
1.328
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr1_-_224993498
|
1.328
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr8_-_6407882
|
1.326
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr12_+_55201854
|
1.314
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr11_-_8849485
|
1.312
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr5_+_61910318
|
1.306
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr12_-_15706340
|
1.302
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr3_+_25444753
|
1.300
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr7_+_838663
|
1.298
|
NM_001130965
|
SUN1
|
Sad1 and UNC84 domain containing 1
|
|
chr18_+_3437607
|
1.298
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr11_+_101488380
|
1.298
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_25445071
|
1.297
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr4_-_149582948
|
1.282
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr6_-_112682573
|
1.269
|
|
LAMA4
|
laminin, alpha 4
|
|
chr2_-_37727472
|
1.266
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr9_-_83493415
|
1.259
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr2_-_192420168
|
1.254
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr9_-_116920232
|
1.237
|
NM_002160
|
TNC
|
tenascin C
|
|
chr15_-_68842799
|
1.236
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr9_-_116608113
|
1.233
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr2_+_108637916
|
1.231
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr5_+_38882017
|
1.228
|
|
OSMR
|
oncostatin M receptor
|
|
chr1_+_24518501
|
1.224
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr10_-_29963906
|
1.219
|
NM_021738
|
SVIL
|
supervillin
|
|
chr2_-_192419896
|
1.215
|
|
SDPR
|
serum deprivation response
|
|
chr3_+_69895651
|
1.214
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_13269562
|
1.201
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr4_+_169654742
|
1.198
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr21_-_26867138
|
1.191
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr1_+_183970093
|
1.188
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr4_-_104240413
|
1.186
|
NM_020139
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2
|
|
chr10_-_62373828
|
1.177
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr4_+_95595419
|
1.174
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr7_+_22733290
|
1.170
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr12_+_50592379
|
1.165
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr1_-_149047329
|
1.159
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr9_-_15240113
|
1.157
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr3_-_150578207
|
1.153
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr12_-_9913724
|
1.149
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr4_+_146820805
|
1.146
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|