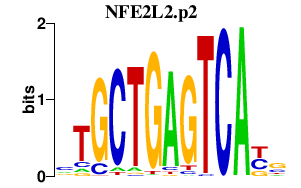
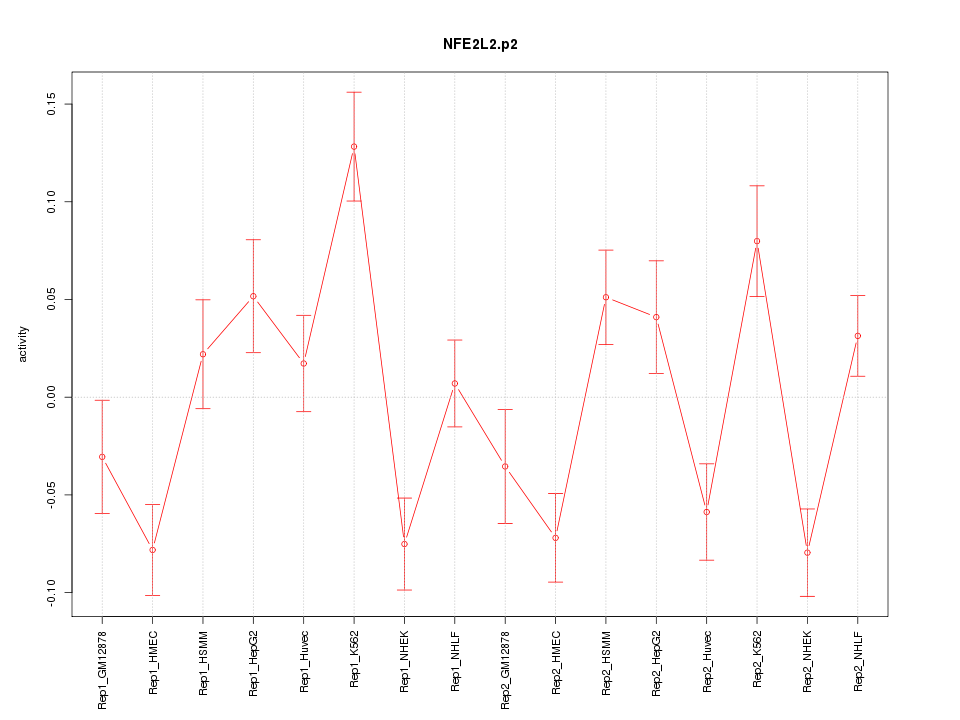
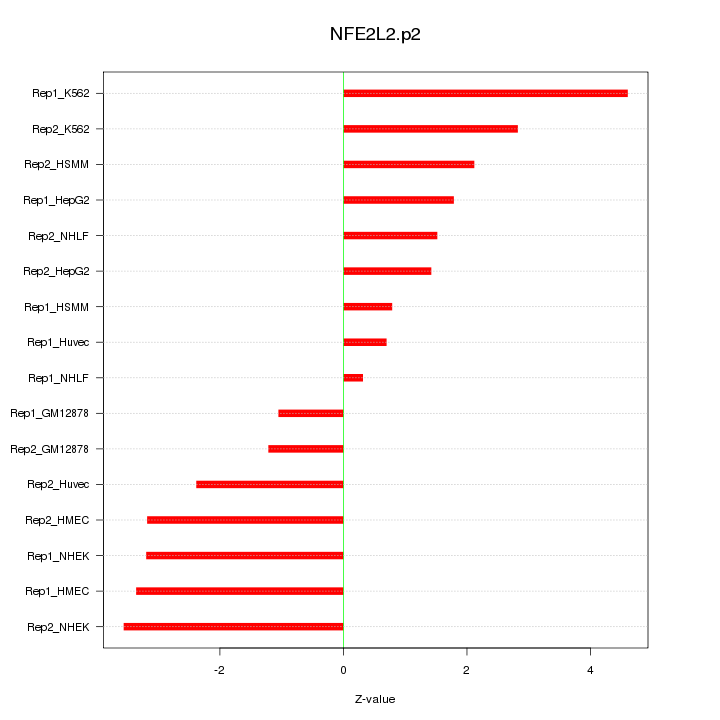
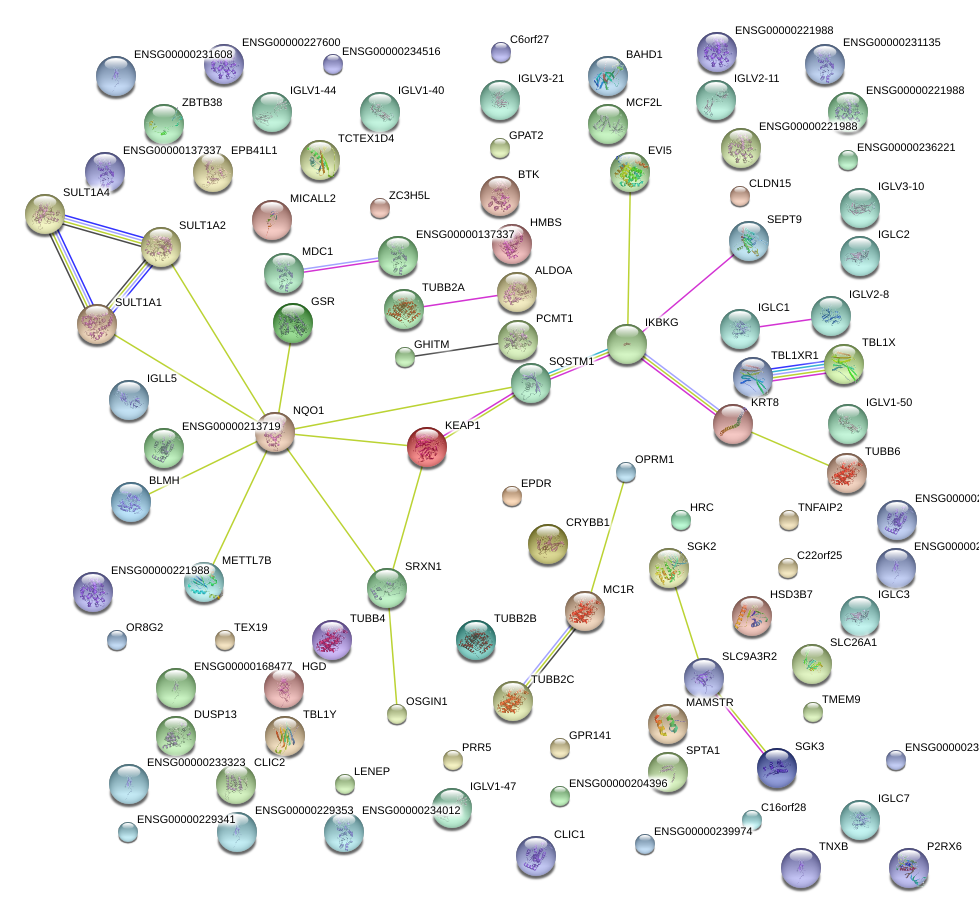
|
chr4_-_977163
|
7.056
|
NM_022042
NM_134425
NM_213613
|
SLC26A1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr16_+_2023279
|
5.359
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr10_-_76538859
|
4.895
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr19_-_54350457
|
4.417
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr20_-_581844
|
4.211
|
NM_080725
|
SRXN1
|
sulfiredoxin 1
|
|
chr22_+_43451600
|
4.144
|
|
PRR5
|
proline rich 5 (renal)
|
|
chr22_+_19699441
|
4.037
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr11_+_118463906
|
3.836
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr1_+_153232685
|
3.727
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr8_-_11242408
|
3.685
|
|
LOC100129129
|
hypothetical LOC100129129
|
|
chr22_+_43451351
|
3.618
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chrX_-_100527799
|
3.597
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr1_-_156923111
|
3.577
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr7_+_37926943
|
3.551
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chrX_-_154216929
|
3.547
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr6_+_150112537
|
3.474
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr8_-_30704762
|
3.451
|
|
GSR
|
glutathione reductase
|
|
chr16_-_1369613
|
3.408
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chrX_+_9391314
|
3.352
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr16_+_88515917
|
3.334
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr16_+_29984611
|
3.317
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_+_32229599
|
3.309
|
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr6_-_32121784
|
3.195
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr16_+_29984544
|
3.181
|
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_+_32229278
|
3.142
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr16_+_29984591
|
3.102
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr2_+_87536174
|
3.050
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr2_+_87536106
|
3.009
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr16_+_29984647
|
3.004
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_+_150112511
|
2.998
|
NM_005389
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr16_+_29984621
|
2.936
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_-_31812049
|
2.902
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr14_+_102672745
|
2.883
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chrX_+_153423652
|
2.846
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr19_-_10474266
|
2.822
|
NM_012289
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr4_+_22608252
|
2.784
|
|
|
|
|
chr1_-_45045543
|
2.781
|
NM_001013632
|
TCTEX1D4
|
Tctex1 domain containing 4
|
|
chr17_-_25642962
|
2.770
|
NM_000386
|
BLMH
|
bleomycin hydrolase
|
|
chr6_-_31812266
|
2.767
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr19_-_53914768
|
2.766
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr6_-_31812251
|
2.719
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr6_-_31812260
|
2.717
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr6_+_150112549
|
2.715
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr6_-_30792830
|
2.682
|
|
MDC1
|
mediator of DNA-damage checkpoint 1
|
|
chr6_-_31812283
|
2.639
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr6_-_31853027
|
2.631
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr12_-_51629874
|
2.623
|
|
KRT8
|
keratin 8
|
|
chr6_-_31812214
|
2.618
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr17_+_72883759
|
2.535
|
NM_001113494
|
SEPT9
|
septin 9
|
|
chr6_-_30793436
|
2.520
|
NM_014641
|
MDC1
|
mediator of DNA-damage checkpoint 1
|
|
chr3_+_142569988
|
2.518
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr17_+_77910411
|
2.482
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr20_+_34206070
|
2.463
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr12_+_54361596
|
2.434
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr16_-_28528149
|
2.429
|
NM_177530
NM_177534
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr6_-_31812297
|
2.396
|
NM_001288
|
CLIC1
|
chloride intracellular channel 1
|
|
chr16_-_28515195
|
2.383
|
NM_177528
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chr6_-_31812072
|
2.382
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr1_-_199390183
|
2.368
|
|
TMEM9
|
transmembrane protein 9
|
|
chr7_-_1465487
|
2.360
|
|
MICALL2
|
MICAL-like 2
|
|
chr5_+_179180447
|
2.356
|
NM_003900
|
SQSTM1
|
sequestosome 1
|
|
chr12_+_54361685
|
2.330
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr16_+_82544327
|
2.264
|
NM_182981
|
OSGIN1
|
oxidative stress induced growth inhibitor 1
|
|
chr22_-_25344037
|
2.264
|
|
CRYBB1
|
crystallin, beta B1
|
|
chr22_-_25343959
|
2.244
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr5_+_179180509
|
2.223
|
|
SQSTM1
|
sequestosome 1
|
|
chr11_+_123600553
|
2.213
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr20_+_41621119
|
2.192
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr16_-_68317945
|
2.152
|
NM_000903
NM_001025433
NM_001025434
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr16_+_30903963
|
2.128
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr20_+_41621079
|
2.103
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr5_+_179180517
|
2.083
|
|
SQSTM1
|
sequestosome 1
|
|
chr7_-_100667750
|
2.080
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr6_+_154449334
|
2.079
|
NM_001145287
|
OPRM1
|
opioid receptor, mu 1
|
|
chr1_-_93030538
|
2.066
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr16_-_28528780
|
2.047
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr22_+_18388656
|
2.031
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr14_-_20642214
|
2.023
|
|
|
|
|
chr22_+_21042091
|
2.021
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr10_+_85889164
|
2.017
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr13_+_112746908
|
2.014
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr15_+_38520594
|
2.014
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr22_+_21079355
|
1.984
|
|
|
|
|
chr3_-_121883647
|
1.945
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr3_-_121884091
|
1.938
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr19_+_18560494
|
1.816
|
NM_001100418
NM_001100419
|
C19orf60
|
chromosome 19 open reading frame 60
|
|
chr2_-_11523666
|
1.798
|
|
E2F6
|
E2F transcription factor 6
|
|
chr22_+_21053972
|
1.795
|
|
|
|
|
chr2_-_219881937
|
1.791
|
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr16_-_30032330
|
1.791
|
NM_024307
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3
|
|
chr11_-_66898223
|
1.788
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr2_-_11523727
|
1.771
|
NM_198256
|
E2F6
|
E2F transcription factor 6
|
|
chr19_-_50977592
|
1.763
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_+_129962292
|
1.756
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr22_+_21495273
|
1.755
|
|
|
|
|
chr22_+_20316968
|
1.755
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr1_-_7923464
|
1.748
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr6_-_28429950
|
1.745
|
NM_145909
|
ZNF323
|
zinc finger protein 323
|
|
chr16_+_30876115
|
1.717
|
NM_014712
|
SETD1A
|
SET domain containing 1A
|
|
chr17_+_44429748
|
1.709
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr17_+_14145051
|
1.707
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr3_-_115826481
|
1.701
|
NM_001164346
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_115880372
|
1.676
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr22_+_21116283
|
1.632
|
|
|
|
|
chr17_-_77474597
|
1.547
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr1_+_19511326
|
1.529
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr22_+_21370394
|
1.529
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr16_+_155972
|
1.525
|
NM_001003938
|
HBM
|
hemoglobin, mu
|
|
chr20_-_581808
|
1.522
|
|
SRXN1
|
sulfiredoxin 1
|
|
chr15_-_32875040
|
1.512
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr12_+_63084317
|
1.512
|
NM_007235
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr22_+_21094094
|
1.505
|
|
|
|
|
chr21_+_31953803
|
1.494
|
NM_000454
|
SOD1
|
superoxide dismutase 1, soluble
|
|
chr9_+_131291224
|
1.448
|
|
LOC100506190
|
hypothetical LOC100506190
|
|
chr7_+_72883128
|
1.445
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr20_+_48240867
|
1.412
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr22_-_17846621
|
1.410
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr22_-_17846711
|
1.403
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr22_-_17846725
|
1.401
|
NM_001035247
NM_005659
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr19_-_50977485
|
1.397
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr17_+_27795635
|
1.389
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr11_+_46358909
|
1.383
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr9_-_115880494
|
1.363
|
NM_001633
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr9_-_115880425
|
1.349
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr17_-_35140307
|
1.332
|
NM_032339
|
C17orf37
|
chromosome 17 open reading frame 37
|
|
chr9_-_126309510
|
1.329
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chrX_-_70243362
|
1.327
|
NM_001025265
|
CXorf65
|
chromosome X open reading frame 65
|
|
chr17_-_53764867
|
1.325
|
|
|
|
|
chr22_+_21407066
|
1.308
|
|
|
|
|
chr1_-_159543847
|
1.303
|
|
MPZ
|
myelin protein zero
|
|
chr19_+_1199548
|
1.290
|
NM_177401
|
MIDN
|
midnolin
|
|
chr3_+_156280129
|
1.289
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_114103616
|
1.288
|
|
PHTF1
|
putative homeodomain transcription factor 1
|
|
chr2_-_224175293
|
1.284
|
|
SCG2
|
secretogranin II
|
|
chr1_-_45249603
|
1.280
|
NM_024602
|
HECTD3
|
HECT domain containing 3
|
|
chr1_-_52117021
|
1.279
|
NM_001101662
NM_002525
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr6_+_64289585
|
1.256
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr5_-_149515613
|
1.246
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr12_-_49897743
|
1.235
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr15_-_81037554
|
1.234
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr22_+_21393107
|
1.231
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr19_-_50977586
|
1.228
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr6_+_32003380
|
1.227
|
|
C2
|
complement component 2
|
|
chr17_+_77529070
|
1.225
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr22_+_20899155
|
1.225
|
|
|
|
|
chr19_+_45564549
|
1.212
|
|
PLD3
|
phospholipase D family, member 3
|
|
chr16_+_30326864
|
1.212
|
|
ZNF771
|
zinc finger protein 771
|
|
chr6_+_31782618
|
1.210
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr1_-_94147574
|
1.209
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr15_+_57691272
|
1.207
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr5_-_149515502
|
1.206
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr9_+_131291144
|
1.206
|
|
|
|
|
chr10_-_71562520
|
1.204
|
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr17_-_35140255
|
1.202
|
|
C17orf37
|
chromosome 17 open reading frame 37
|
|
chr6_+_36518521
|
1.198
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr1_-_176205672
|
1.197
|
NM_033127
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr9_-_115880366
|
1.185
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chrX_-_152863251
|
1.183
|
NM_002910
|
RENBP
|
renin binding protein
|
|
chr16_-_73830480
|
1.174
|
NM_001170721
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr16_+_72888231
|
1.171
|
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr1_-_52116965
|
1.169
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr12_+_54910767
|
1.169
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr1_-_52116994
|
1.167
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr7_+_150319076
|
1.158
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr22_-_39189369
|
1.157
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr16_+_72888161
|
1.155
|
NM_002811
|
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr10_+_121400926
|
1.154
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr16_+_28413421
|
1.146
|
NM_018690
|
APOBR
|
apolipoprotein B receptor
|
|
chr5_-_95184221
|
1.143
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr6_-_36518643
|
1.140
|
NM_152990
|
PXT1
|
peroxisomal, testis specific 1
|
|
chr6_-_101018271
|
1.138
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr1_-_94147616
|
1.131
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr3_-_39209075
|
1.113
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr1_-_172258129
|
1.112
|
|
|
|
|
chr15_+_57691382
|
1.105
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr6_+_36518778
|
1.099
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr2_+_119905812
|
1.094
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr10_+_121400881
|
1.090
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr8_-_59734975
|
1.090
|
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr16_+_30326386
|
1.089
|
|
ZNF771
|
zinc finger protein 771
|
|
chr19_-_56260135
|
1.087
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr3_-_180460372
|
1.085
|
NM_171829
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr10_+_121400741
|
1.069
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|
|
chr8_-_59734939
|
1.065
|
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr16_+_30326077
|
1.063
|
NM_001142305
NM_016643
|
ZNF771
|
zinc finger protein 771
|
|
chr3_-_195571748
|
1.060
|
NM_001135057
NM_130830
|
LRRC15
|
leucine rich repeat containing 15
|
|
chrX_-_110400406
|
1.060
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr11_+_2881080
|
1.058
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr10_+_134751398
|
1.054
|
NM_001083909
|
GPR123
|
G protein-coupled receptor 123
|
|
chr8_-_59734957
|
1.048
|
NM_003580
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr1_-_223683141
|
1.046
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr1_-_109770474
|
1.035
|
NM_002790
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr14_+_38772870
|
1.034
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr22_+_25347927
|
1.031
|
NM_001886
|
CRYBA4
|
crystallin, beta A4
|
|
chr20_+_48240705
|
1.028
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr1_-_6585485
|
1.025
|
NM_014851
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr10_-_71562602
|
1.021
|
NM_001198696
NM_032797
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2
|
|
chr5_-_95184121
|
1.008
|
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr12_-_54387732
|
1.008
|
|
ITGA7
|
integrin, alpha 7
|
|
chr1_-_45249554
|
1.004
|
|
HECTD3
|
HECT domain containing 3
|