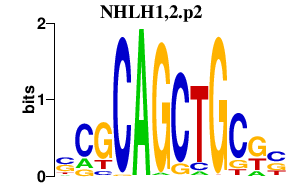
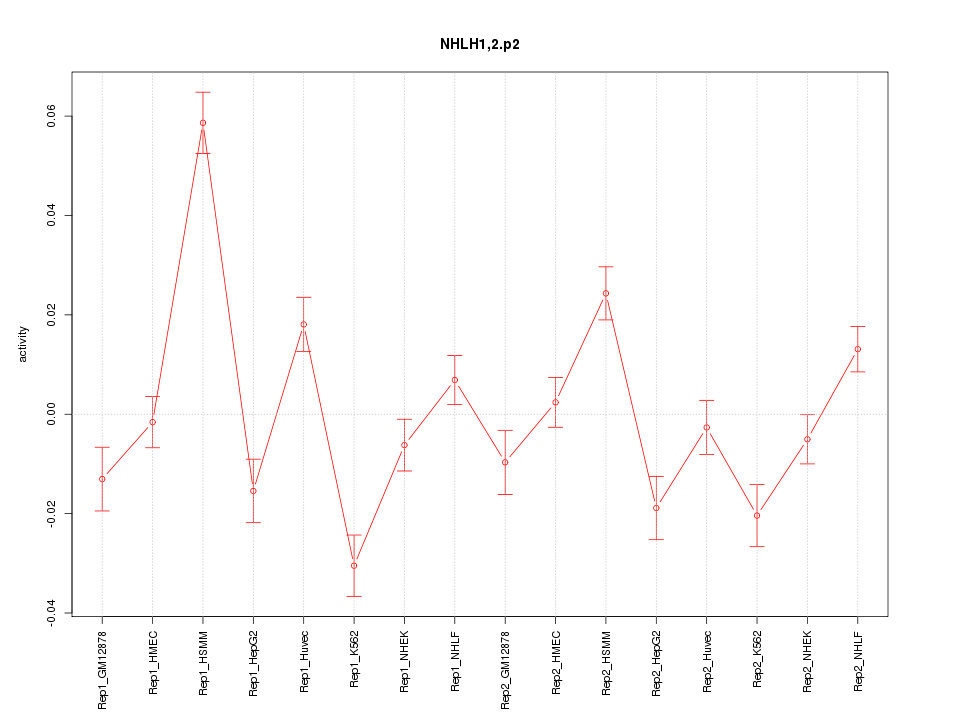
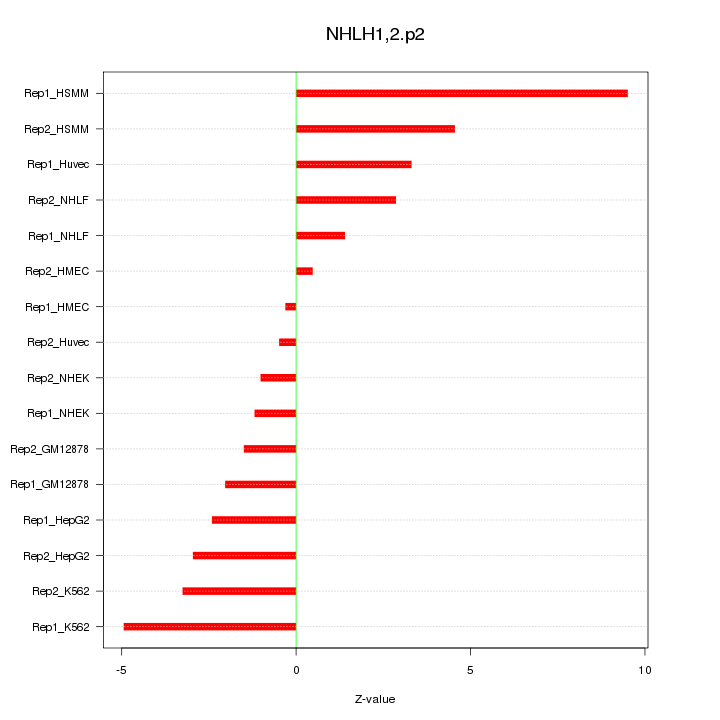
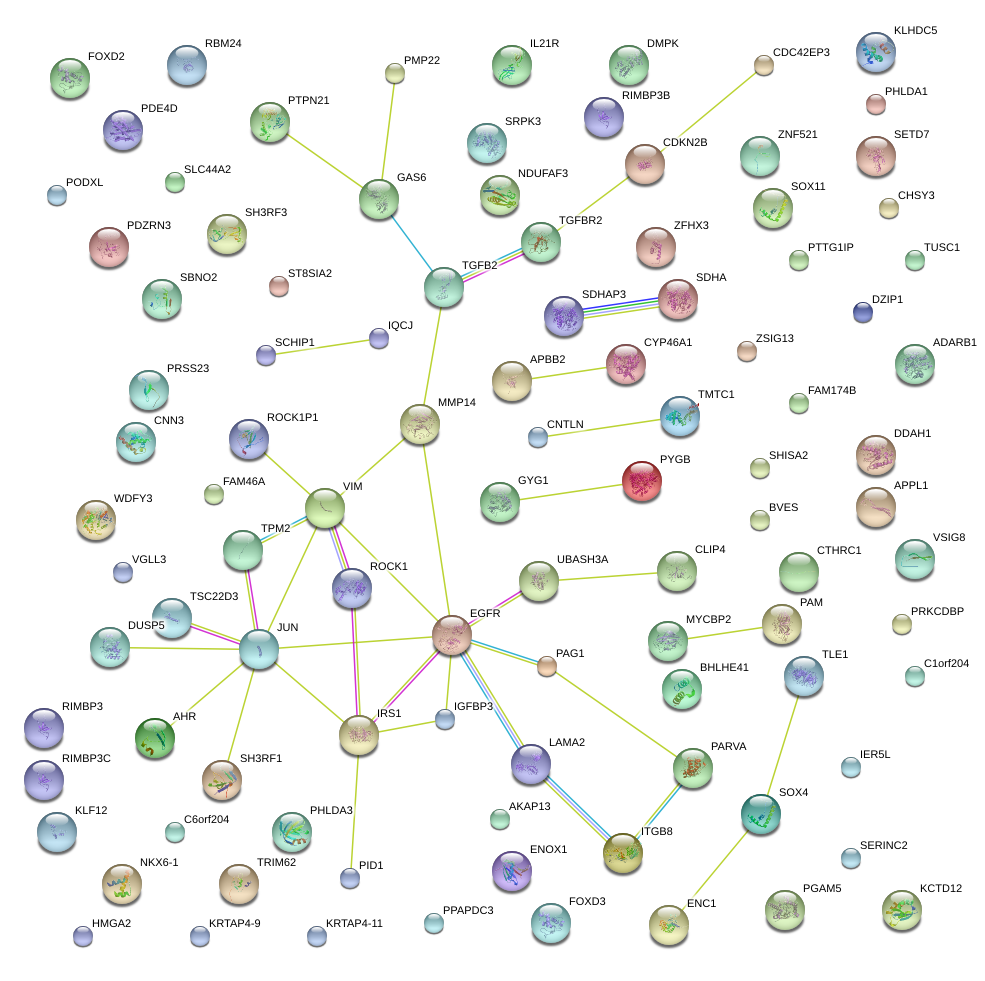
|
chr2_+_109112428
|
6.156
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr7_+_17304707
|
6.007
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr13_-_25523164
|
4.927
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr11_-_6298284
|
4.761
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr4_-_40911212
|
4.669
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr9_+_17124974
|
3.976
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr22_+_20068039
|
3.805
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr14_-_88090641
|
3.785
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr1_-_33420213
|
3.616
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr12_-_74710741
|
3.497
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr2_-_229844198
|
3.383
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr12_+_27824429
|
3.350
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr21_+_45318861
|
3.226
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr3_-_73756668
|
3.162
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr3_-_87122936
|
3.086
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr9_-_83493415
|
3.079
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr2_-_37752731
|
3.036
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr22_-_20235372
|
3.020
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr3_-_87122958
|
3.005
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr5_-_58370693
|
2.928
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_-_71650034
|
2.917
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chrX_+_152699616
|
2.916
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr1_-_59020875
|
2.910
|
|
JUN
|
jun proto-oncogene
|
|
chr19_+_10597167
|
2.908
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr5_-_72779585
|
2.900
|
|
FOXD1
|
forkhead box D1
|
|
chr10_+_112247614
|
2.870
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr19_-_1083109
|
2.862
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr9_-_21549696
|
2.795
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr2_-_227371698
|
2.776
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr6_-_82518576
|
2.720
|
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr9_-_35679799
|
2.712
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chrX_-_106846253
|
2.671
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr9_-_35679902
|
2.663
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr4_-_86106567
|
2.660
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr17_+_36515166
|
2.656
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr14_+_99220349
|
2.602
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr1_+_68070573
|
2.566
|
|
|
|
|
chr6_-_119079712
|
2.557
|
NM_001042475
NM_206921
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr1_-_158091296
|
2.537
|
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr19_-_50977485
|
2.530
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr10_+_17311307
|
2.517
|
|
VIM
|
vimentin
|
|
chr13_-_43259032
|
2.516
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr6_+_3696224
|
2.516
|
|
|
|
|
chr2_+_29191711
|
2.514
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr14_+_22375753
|
2.500
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr7_-_45927263
|
2.498
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr6_-_82519093
|
2.483
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr12_-_26166485
|
2.464
|
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr3_+_49034050
|
2.459
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr1_+_31658549
|
2.454
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr10_+_17311283
|
2.449
|
|
VIM
|
vimentin
|
|
chr13_-_73606362
|
2.437
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr6_-_105691675
|
2.434
|
|
BVES
|
blood vessel epicardial substance
|
|
chr15_+_90738108
|
2.430
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr10_+_17311242
|
2.414
|
|
VIM
|
vimentin
|
|
chr8_-_82186832
|
2.406
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr20_+_25176698
|
2.403
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr10_+_17311282
|
2.394
|
|
VIM
|
vimentin
|
|
chr13_-_76358370
|
2.386
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr10_+_99463446
|
2.384
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr13_-_95094920
|
2.356
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr10_+_17311347
|
2.329
|
|
VIM
|
vimentin
|
|
chr9_+_133154896
|
2.317
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr17_-_36528113
|
2.313
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr7_+_55054416
|
2.311
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr10_+_99463468
|
2.309
|
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr1_-_85703321
|
2.301
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_-_140696740
|
2.269
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr12_+_64504835
|
2.264
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr13_+_113546841
|
2.250
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr6_-_105691235
|
2.248
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr11_+_12355600
|
2.179
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr6_+_129245978
|
2.177
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr1_-_199704773
|
2.124
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr15_+_83724743
|
2.119
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr9_-_21999270
|
2.107
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr17_-_15106579
|
2.104
|
|
PMP22
|
peripheral myelin protein 22
|
|
chr12_-_29827952
|
2.091
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr5_-_73972055
|
2.091
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr11_+_86189138
|
2.084
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr9_-_130980359
|
2.083
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr7_-_130891861
|
2.081
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr7_+_20336849
|
2.073
|
|
ITGB8
|
integrin, beta 8
|
|
chr18_-_21185968
|
2.059
|
|
ZNF521
|
zinc finger protein 521
|
|
chr1_+_216586199
|
2.055
|
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr21_-_45118037
|
2.055
|
NM_004339
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr3_+_57236818
|
2.054
|
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr3_+_160964574
|
2.053
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_15106597
|
2.051
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_95165089
|
2.040
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr12_+_27824211
|
2.033
|
|
KLHDC5
|
kelch domain containing 5
|
|
chr21_+_45118353
|
2.026
|
|
|
|
|
chr18_-_16945613
|
2.016
|
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr16_+_27320983
|
2.005
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr9_-_25668230
|
1.989
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr3_+_150191884
|
1.976
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr5_+_129268352
|
1.968
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr15_-_90999606
|
1.962
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr13_+_113546897
|
1.943
|
|
GAS6
|
growth arrest-specific 6
|
|
chr18_-_16945763
|
1.941
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr5_+_102229612
|
1.932
|
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr21_-_45117989
|
1.923
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr13_+_113546943
|
1.918
|
|
GAS6
|
growth arrest-specific 6
|
|
chr21_-_45118026
|
1.914
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr8_+_104452877
|
1.892
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr5_+_271355
|
1.866
|
NM_004168
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr3_+_30623376
|
1.859
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr2_+_5750229
|
1.856
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_+_17389752
|
1.856
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr14_+_58174489
|
1.833
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr13_-_73605914
|
1.829
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr1_-_84237316
|
1.827
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr11_+_110916629
|
1.816
|
|
LAYN
|
layilin
|
|
chr3_-_185912240
|
1.808
|
NM_022149
|
MAGEF1
|
melanoma antigen family F, 1
|
|
chr2_-_19421852
|
1.789
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr5_+_102229425
|
1.777
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr11_+_133444175
|
1.754
|
|
JAM3
|
junctional adhesion molecule 3
|
|
chr8_+_104452961
|
1.750
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr12_+_51730101
|
1.743
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr3_-_147361465
|
1.731
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr4_-_57671307
|
1.725
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr19_-_50977586
|
1.716
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr10_+_27833167
|
1.709
|
NM_021252
|
RAB18
|
RAB18, member RAS oncogene family
|
|
chr21_-_45117960
|
1.693
|
|
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein
|
|
chr3_-_151171576
|
1.691
|
|
PFN2
|
profilin 2
|
|
chr3_+_49034494
|
1.685
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr1_+_15145001
|
1.681
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr9_+_101623957
|
1.681
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr11_+_44543716
|
1.679
|
NM_001024844
NM_002231
|
CD82
|
CD82 molecule
|
|
chr3_+_151171755
|
1.674
|
|
LOC646903
|
hypothetical LOC646903
|
|
chr16_+_82490230
|
1.667
|
NM_012213
|
MLYCD
|
malonyl-CoA decarboxylase
|
|
chr1_+_238321794
|
1.665
|
NM_020066
|
FMN2
|
formin 2
|
|
chr17_+_36493927
|
1.660
|
NM_033061
|
KRTAP4-7
|
keratin associated protein 4-7
|
|
chr14_-_60185659
|
1.653
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr2_-_219404755
|
1.647
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr10_+_104667991
|
1.645
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr5_+_271428
|
1.645
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr20_-_23566325
|
1.644
|
NM_000099
|
CST3
|
cystatin C
|
|
chr6_-_33864863
|
1.636
|
|
LEMD2
|
LEM domain containing 2
|
|
chr9_+_109085372
|
1.636
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr6_-_33864876
|
1.632
|
|
LEMD2
|
LEM domain containing 2
|
|
chr9_+_109085337
|
1.630
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr19_-_50977592
|
1.629
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr5_+_271447
|
1.625
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr16_+_53522611
|
1.623
|
NM_005853
|
IRX5
|
iroquois homeobox 5
|
|
chr1_-_22136263
|
1.621
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr2_+_56264627
|
1.620
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr5_+_72148173
|
1.605
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr18_+_19848749
|
1.605
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr7_-_526005
|
1.601
|
NM_002607
NM_033023
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr10_+_72642274
|
1.589
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr1_-_143750988
|
1.580
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr9_-_71476917
|
1.579
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr9_+_71848555
|
1.578
|
|
MAMDC2
|
MAM domain containing 2
|
|
chrX_+_73557809
|
1.574
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr2_+_202949289
|
1.569
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr3_-_121550767
|
1.565
|
NM_001099678
|
LRRC58
|
leucine rich repeat containing 58
|
|
chr2_+_191818124
|
1.565
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr17_-_53387493
|
1.558
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr7_+_129919330
|
1.557
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr3_-_126088530
|
1.552
|
|
ITGB5
|
integrin, beta 5
|
|
chr12_+_62523648
|
1.546
|
|
|
|
|
chr9_+_98252200
|
1.543
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr6_-_128883125
|
1.542
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr17_+_1906262
|
1.525
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr15_+_72006015
|
1.524
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr1_-_72520735
|
1.513
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr19_-_2207415
|
1.511
|
NM_144616
|
JSRP1
|
junctional sarcoplasmic reticulum protein 1
|
|
chr1_-_154737162
|
1.511
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr11_+_7997366
|
1.491
|
|
TUB
|
tubby homolog (mouse)
|
|
chr2_+_241586927
|
1.485
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr13_-_22905822
|
1.484
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr10_-_125841840
|
1.475
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr10_+_124211353
|
1.473
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr5_-_54317102
|
1.471
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr3_+_57236816
|
1.466
|
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr16_-_3295375
|
1.465
|
|
|
|
|
chr19_-_63301402
|
1.456
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr2_+_202607554
|
1.455
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr12_-_50871953
|
1.446
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr4_-_57671273
|
1.446
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr4_-_81213207
|
1.442
|
|
|
|
|
chr18_+_19848347
|
1.441
|
NM_001135993
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr11_+_124539814
|
1.438
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr1_-_39877928
|
1.429
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr2_+_56265260
|
1.423
|
|
|
|
|
chr4_-_122837579
|
1.418
|
|
ANXA5
|
annexin A5
|
|
chr3_+_57236769
|
1.414
|
NM_012096
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr10_-_62373828
|
1.409
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr8_+_98856984
|
1.409
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr4_+_86615275
|
1.407
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr5_+_14196287
|
1.406
|
NM_007118
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr20_+_773283
|
1.404
|
NM_031424
|
FAM110A
|
family with sequence similarity 110, member A
|
|
chr7_+_98084531
|
1.400
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr11_+_124539768
|
1.398
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr1_-_154737189
|
1.394
|
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr7_+_27102508
|
1.391
|
|
|
|
|
chr2_+_85834111
|
1.385
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr19_-_63301386
|
1.365
|
|
ZSCAN18
|
zinc finger and SCAN domain containing 18
|
|
chr6_-_33864883
|
1.363
|
NM_181336
|
LEMD2
|
LEM domain containing 2
|