|
chr2_+_171280046
|
2.318
|
NM_001003845
|
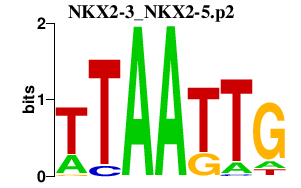
SP5
|
Sp5 transcription factor
|
|
chr2_+_171280182
|
2.219
|
|
|
|
|
chr4_+_90252990
|
1.821
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr3_-_121884091
|
1.520
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr7_+_94135384
|
1.484
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_53849888
|
1.471
|
|
RNF43
|
ring finger protein 43
|
|
chr6_+_105511615
|
1.380
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr3_-_121883647
|
1.239
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr12_-_23995109
|
1.217
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr10_+_7785360
|
1.127
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr12_-_23995220
|
1.116
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chrX_+_115481774
|
1.113
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr11_-_16380967
|
1.111
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_+_105823723
|
1.051
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr10_+_7785347
|
1.050
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr9_+_129066576
|
1.008
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr10_-_73953699
|
1.002
|
NM_001195519
|
CBARA1
|
calcium binding atopy-related autoantigen 1
|
|
chrX_-_24600661
|
0.994
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr10_+_7785290
|
0.948
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr17_-_61655915
|
0.941
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr10_+_7785331
|
0.933
|
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr11_-_18214891
|
0.905
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr20_+_5935897
|
0.900
|
NM_001127458
|
CRLS1
|
cardiolipin synthase 1
|
|
chr13_+_23042722
|
0.891
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr9_-_14858974
|
0.865
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr4_+_69716298
|
0.847
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr17_+_24394281
|
0.819
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr4_+_74493174
|
0.812
|
|
ALB
|
albumin
|
|
chr4_-_69852090
|
0.812
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr7_+_113842284
|
0.799
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr16_+_7322751
|
0.757
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr8_-_72436519
|
0.756
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr9_-_116305273
|
0.752
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr19_+_50100843
|
0.740
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr17_-_46479127
|
0.728
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_+_53787187
|
0.725
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr4_+_74520796
|
0.724
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr7_+_116238309
|
0.721
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr14_+_31868229
|
0.701
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr12_-_16652202
|
0.683
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_74493301
|
0.680
|
|
ALB
|
albumin
|
|
chr19_-_53081383
|
0.679
|
NM_003167
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr3_+_89239494
|
0.673
|
|
EPHA3
|
EPH receptor A3
|
|
chr8_-_72437020
|
0.672
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_-_165133239
|
0.655
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr8_-_72436832
|
0.652
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chrX_-_129347101
|
0.645
|
NM_178471
|
GPR119
|
G protein-coupled receptor 119
|
|
chr10_-_52315390
|
0.643
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr16_+_20682812
|
0.643
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr6_+_50789215
|
0.642
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr19_-_53081326
|
0.639
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr4_+_74493266
|
0.636
|
|
ALB
|
albumin
|
|
chr5_+_59819761
|
0.632
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr17_-_36346240
|
0.622
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr11_+_101488380
|
0.621
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr10_+_24795446
|
0.619
|
|
KIAA1217
|
KIAA1217
|
|
chr10_+_101532544
|
0.616
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chrX_+_115481848
|
0.613
|
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr10_+_7785235
|
0.605
|
NM_002216
|
ITIH2
|
inter-alpha (globulin) inhibitor H2
|
|
chr17_-_36418869
|
0.597
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chrX_-_48212683
|
0.595
|
|
SLC38A5
|
solute carrier family 38, member 5
|
|
chr4_+_74493218
|
0.588
|
|
ALB
|
albumin
|
|
chr10_+_24795465
|
0.588
|
|
KIAA1217
|
KIAA1217
|
|
chr4_-_176970895
|
0.586
|
|
GPM6A
|
glycoprotein M6A
|
|
chr5_-_131907071
|
0.584
|
NM_000879
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil)
|
|
chr10_-_52315436
|
0.564
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr12_-_16650697
|
0.564
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16650687
|
0.559
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_+_74493331
|
0.541
|
|
ALB
|
albumin
|
|
chr13_-_35327798
|
0.536
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr3_+_46894239
|
0.533
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr10_+_101532450
|
0.532
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr17_-_1195096
|
0.530
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr6_-_26038817
|
0.523
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr10_+_102722275
|
0.520
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chrX_+_68752635
|
0.514
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr4_-_23500706
|
0.508
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr5_-_59819642
|
0.504
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr14_-_37134050
|
0.496
|
|
FOXA1
|
forkhead box A1
|
|
chr1_+_50342168
|
0.487
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr13_+_77213295
|
0.478
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr6_-_46401174
|
0.473
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_+_28237529
|
0.468
|
NM_001145129
|
ZNF389
|
zinc finger protein 389
|
|
chr4_+_74499651
|
0.468
|
|
ALB
|
albumin
|
|
chr3_+_160269836
|
0.467
|
|
IQCJ
|
IQ motif containing J
|
|
chr4_-_186970366
|
0.458
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_31731647
|
0.450
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr12_+_49605010
|
0.450
|
|
METTL7A
|
methyltransferase like 7A
|
|
chr17_-_36456949
|
0.441
|
NM_001123387
|
KRTAP2-1
|
keratin associated protein 2-1
|
|
chr11_+_86710076
|
0.439
|
|
TMEM135
|
transmembrane protein 135
|
|
chr20_+_30160952
|
0.439
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr8_+_9990471
|
0.437
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr2_-_200033445
|
0.436
|
|
SATB2
|
SATB homeobox 2
|
|
chr22_-_16127520
|
0.435
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr2_-_215953818
|
0.432
|
|
FN1
|
fibronectin 1
|
|
chr17_-_36347197
|
0.430
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chrX_+_90976314
|
0.422
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr16_+_70654625
|
0.422
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr13_-_93929719
|
0.421
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr14_-_37134191
|
0.420
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chrX_-_135783496
|
0.416
|
|
|
|
|
chr1_+_556968
|
0.412
|
|
|
|
|
chr12_+_54700955
|
0.410
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr6_+_31731667
|
0.410
|
|
APOM
|
apolipoprotein M
|
|
chr1_-_214963279
|
0.407
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr2_+_189547377
|
0.405
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr11_-_16454493
|
0.402
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_+_225973845
|
0.399
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr12_+_50342814
|
0.398
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chrX_+_9391314
|
0.396
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr2_+_161981046
|
0.396
|
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_36991189
|
0.393
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr6_+_155579788
|
0.393
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chrX_-_106033202
|
0.388
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chrX_+_21960841
|
0.386
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr3_+_160269734
|
0.385
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chrX_+_123062143
|
0.382
|
|
STAG2
|
stromal antigen 2
|
|
chr8_-_82536312
|
0.381
|
NM_001080526
|
FABP9
|
fatty acid binding protein 9, testis
|
|
chr2_+_189547358
|
0.381
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr18_+_71051713
|
0.377
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr2_+_161981138
|
0.376
|
|
TBR1
|
T-box, brain, 1
|
|
chr7_-_96471544
|
0.369
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr14_+_69307914
|
0.369
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr6_-_24491498
|
0.366
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr12_-_9123126
|
0.365
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr13_+_97856148
|
0.363
|
|
|
|
|
chr12_+_40117751
|
0.360
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr4_-_83566045
|
0.355
|
|
|
|
|
chr6_-_32265489
|
0.355
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr14_-_37133927
|
0.352
|
|
FOXA1
|
forkhead box A1
|
|
chr6_+_41128917
|
0.345
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr2_-_190635566
|
0.345
|
NM_005259
|
MSTN
|
myostatin
|
|
chr11_-_57275828
|
0.340
|
NM_001145101
|
BTBD18
|
BTB (POZ) domain containing 18
|
|
chr1_+_117098579
|
0.339
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chrX_+_70682204
|
0.338
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr12_-_4358968
|
0.337
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr5_+_140751666
|
0.336
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr2_+_161980865
|
0.336
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr9_+_4829763
|
0.334
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr3_-_188492163
|
0.331
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr12_+_49604782
|
0.331
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chr12_-_120725210
|
0.328
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr5_-_135259403
|
0.328
|
NM_000590
|
IL9
|
interleukin 9
|
|
chr4_+_100714995
|
0.328
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr15_-_53996597
|
0.322
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr11_+_89624017
|
0.322
|
|
|
|
|
chr15_+_38082041
|
0.321
|
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr7_+_142053693
|
0.320
|
NM_001190487
|
MTRNR2L6
|
MT-RNR2-like 6
|
|
chr19_-_5789740
|
0.320
|
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr14_+_38772870
|
0.320
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr14_-_64479198
|
0.315
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr2_+_189547323
|
0.314
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr19_-_47724365
|
0.313
|
NM_001024912
NM_001184813
NM_001184815
NM_001184816
NM_001712
|
CEACAM1
|
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein)
|
|
chr10_-_97190885
|
0.313
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr4_+_88939725
|
0.312
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr3_-_15538164
|
0.310
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr16_-_4792857
|
0.309
|
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr3_+_85858321
|
0.307
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr16_+_70646008
|
0.306
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr6_+_132915522
|
0.305
|
NM_053278
|
TAAR8
|
trace amine associated receptor 8
|
|
chr2_+_96818048
|
0.305
|
|
CNNM4
|
cyclin M4
|
|
chr6_-_53121540
|
0.305
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr7_-_112513379
|
0.302
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_+_100089118
|
0.295
|
NM_000646
|
AGL
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
|
|
chr16_-_68317855
|
0.295
|
|
NQO1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr4_-_176971175
|
0.295
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr15_+_56511466
|
0.294
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr8_-_17624009
|
0.292
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr10_+_24537725
|
0.290
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr1_+_50348052
|
0.290
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_+_165804157
|
0.289
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr4_+_140806536
|
0.289
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr3_+_46370578
|
0.288
|
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr2_-_216654708
|
0.288
|
NM_018441
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase
|
|
chr19_+_4442610
|
0.287
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr4_+_71418886
|
0.284
|
NM_212557
|
AMTN
|
amelotin
|
|
chr4_-_100359353
|
0.284
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chrX_-_130251068
|
0.282
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr3_+_162697289
|
0.281
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr8_-_82769761
|
0.280
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr5_-_149649593
|
0.275
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr11_+_91724909
|
0.274
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chrX_-_18600108
|
0.274
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr12_-_16649211
|
0.273
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_-_49712483
|
0.272
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr9_-_103185621
|
0.271
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr7_-_44547382
|
0.271
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr14_+_99555471
|
0.270
|
|
EVL
|
Enah/Vasp-like
|
|
chr20_-_19984685
|
0.269
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr11_+_20577521
|
0.269
|
NM_004211
|
SLC6A5
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 5
|
|
chr22_-_36836620
|
0.267
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr13_+_34948977
|
0.267
|
|
NBEA
|
neurobeachin
|
|
chr4_-_123761615
|
0.264
|
NM_021803
|
IL21
|
interleukin 21
|
|
chr12_-_23993830
|
0.263
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr7_+_147918589
|
0.262
|
NM_145304
|
C7orf33
|
chromosome 7 open reading frame 33
|
|
chr17_-_31352720
|
0.261
|
|
CCL14-CCL15
|
CCL14-CCL15 read-through transcript
|
|
chr5_-_148738980
|
0.261
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr6_+_131190237
|
0.261
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr4_+_123380569
|
0.259
|
|
KIAA1109
|
KIAA1109
|
|
chr6_+_167624792
|
0.258
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|