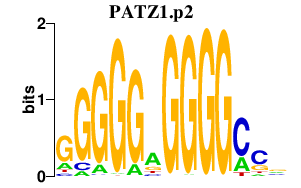
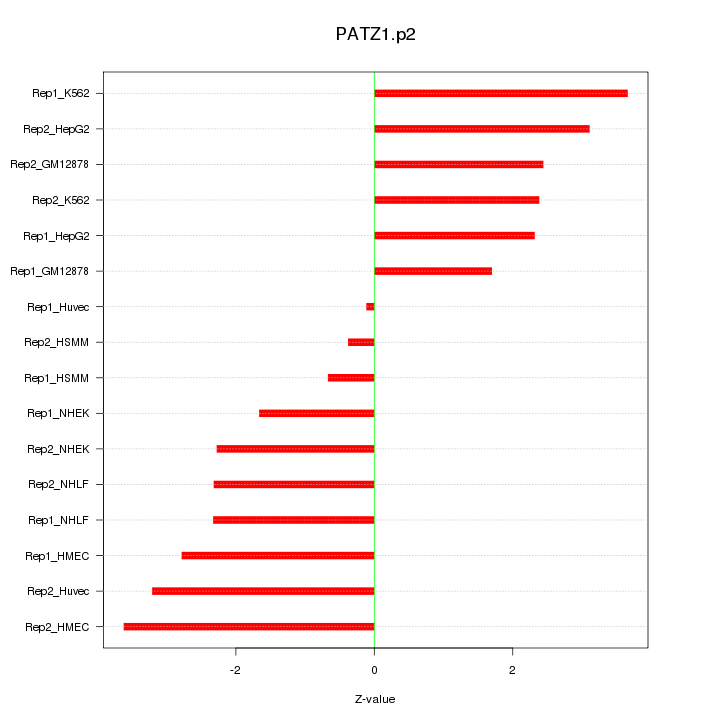
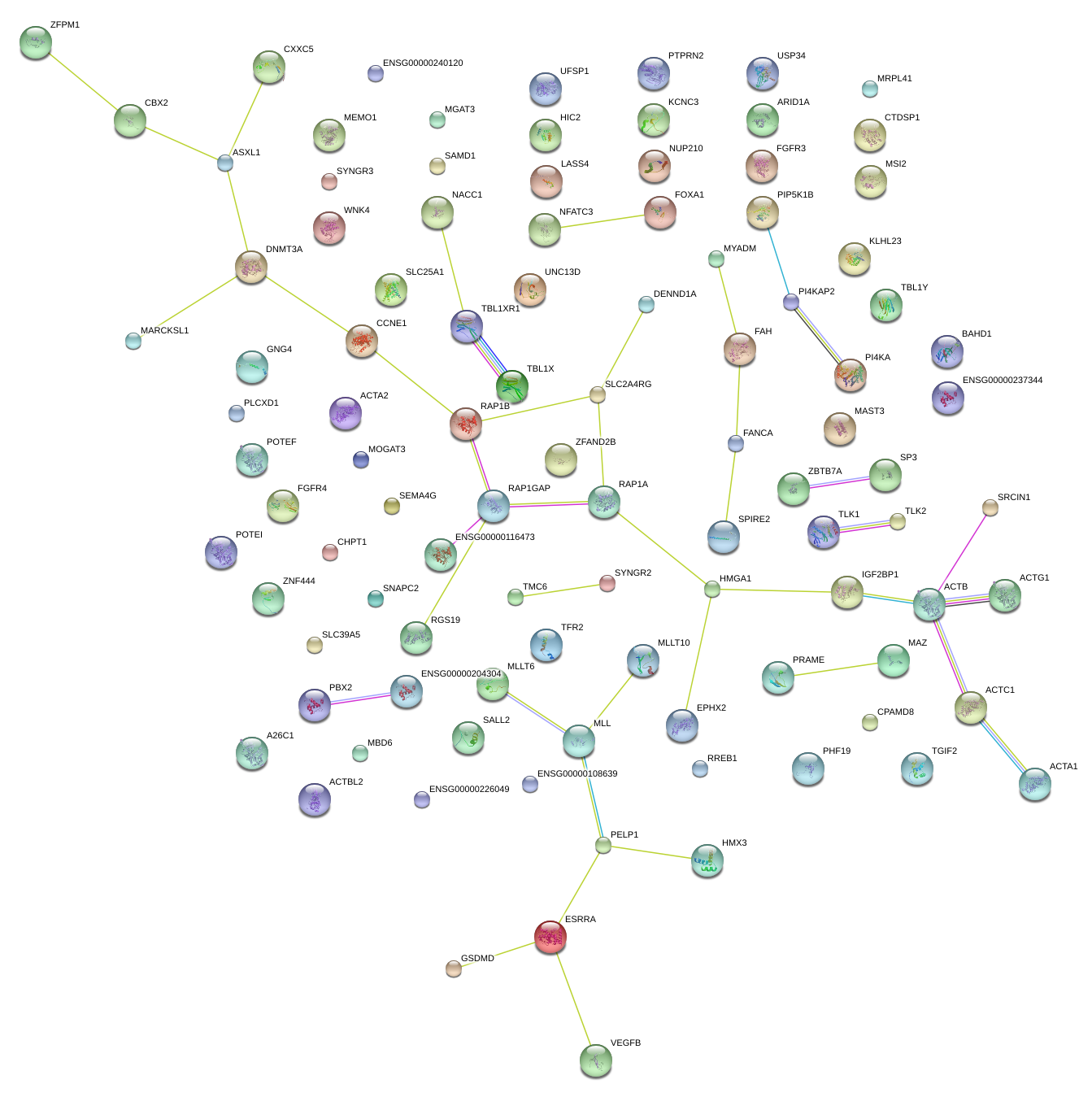
|
chr17_+_34114924
|
8.379
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr17_+_34115383
|
4.023
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_-_25328452
|
3.813
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr11_+_63829616
|
3.666
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr1_-_32574185
|
3.614
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr17_-_73636386
|
3.597
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr17_-_73636362
|
3.502
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr19_-_4017739
|
3.408
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr8_+_144706699
|
3.145
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chrX_+_138060
|
3.089
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr9_-_125732005
|
3.061
|
NM_020946
NM_024820
|
DENND1A
|
DENN/MADD domain containing 1A
|
|
chrX_+_9392980
|
2.894
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_+_3679168
|
2.799
|
NM_001163724
|
LOC388588
|
hypothetical protein LOC388588
|
|
chr19_-_55524445
|
2.676
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr19_+_61344501
|
2.671
|
|
ZNF444
|
zinc finger protein 444
|
|
chr16_+_29725311
|
2.654
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr16_+_87047159
|
2.616
|
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr6_+_34313007
|
2.539
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_+_100615530
|
2.484
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr6_+_34312979
|
2.458
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_+_8180179
|
2.457
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr7_-_100077072
|
2.448
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr19_+_34994701
|
2.410
|
NM_001238
|
CCNE1
|
cyclin E1
|
|
chr19_+_61344341
|
2.393
|
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr9_+_70509925
|
2.384
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr10_+_124885556
|
2.383
|
NM_001105574
|
HMX3
|
H6 family homeobox 3
|
|
chr14_-_21064182
|
2.356
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr22_+_20101661
|
2.354
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr17_+_52688842
|
2.341
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr2_+_170298700
|
2.307
|
|
KLHL23
|
kelch-like 23 (Drosophila)
|
|
chr5_+_176446425
|
2.271
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr9_-_122679387
|
2.270
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr20_+_34635362
|
2.264
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr22_-_18807805
|
2.260
|
|
PI4KAP2
PI4KAP1
|
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 2
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 1
|
|
chr17_-_71352090
|
2.233
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr17_+_44429748
|
2.212
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr20_+_61841662
|
2.210
|
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr17_-_34015631
|
2.207
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr11_+_117812282
|
2.164
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr19_+_7891193
|
2.136
|
NM_003083
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa
|
|
chr10_+_102719239
|
2.065
|
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr1_-_21868380
|
2.063
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr6_+_7052798
|
2.050
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr7_-_100325209
|
2.043
|
NM_001015072
|
UFSP1
|
UFM1-specific peptidase 1 (non-functional)
|
|
chr22_-_17545872
|
2.040
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr17_+_73676219
|
2.033
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr6_+_34313347
|
2.019
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr2_-_171725150
|
2.019
|
|
TLK1
|
tousled-like kinase 1
|
|
chr3_-_13436725
|
2.010
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr16_+_29725597
|
2.000
|
|
|
|
|
chr17_-_71352017
|
2.000
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr14_-_37134191
|
2.000
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr2_+_170298487
|
1.992
|
|
|
|
|
chr2_-_32089608
|
1.986
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr22_-_17546296
|
1.984
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr2_-_32089557
|
1.977
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr6_-_32265489
|
1.969
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr11_+_63758550
|
1.941
|
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr2_+_218972963
|
1.904
|
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr19_+_13090101
|
1.904
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr12_+_56202874
|
1.902
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr22_-_17546234
|
1.897
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr20_+_61841553
|
1.889
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr5_+_139008667
|
1.871
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr3_-_13436802
|
1.855
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr16_+_1979959
|
1.854
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr19_-_14062230
|
1.848
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr19_+_18069596
|
1.847
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chr15_+_78232184
|
1.821
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr22_-_21231421
|
1.811
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr9_+_139566129
|
1.805
|
NM_032477
|
MRPL41
|
mitochondrial ribosomal protein L41
|
|
chr17_+_75366524
|
1.796
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr16_+_88422354
|
1.777
|
NM_032451
|
SPIRE2
|
spire homolog 2 (Drosophila)
|
|
chr12_+_54910767
|
1.765
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr19_+_59064795
|
1.759
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr2_-_61551403
|
1.757
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr1_+_26895108
|
1.757
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr2_-_174537354
|
1.747
|
|
SP3
|
Sp3 transcription factor
|
|
chr8_+_27404508
|
1.727
|
NM_001979
|
EPHX2
|
epoxide hydrolase 2, cytoplasmic
|
|
chr22_-_17546276
|
1.702
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr7_-_158073121
|
1.693
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr17_-_71352182
|
1.688
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr2_+_219779737
|
1.678
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr19_-_16868674
|
1.675
|
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8
|
|
chr1_-_233879617
|
1.673
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr17_+_38186141
|
1.672
|
NM_032387
|
WNK4
|
WNK lysine deficient protein kinase 4
|
|
chr20_+_30409649
|
1.672
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr4_+_1764802
|
1.661
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr15_+_38520594
|
1.660
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr15_+_78232232
|
1.651
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr22_+_38183480
|
1.649
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr1_+_111963920
|
1.633
|
NM_001010935
NM_002884
|
RAP1A
|
RAP1A, member of RAS oncogene family
|
|
chr17_-_77094426
|
1.625
|
|
ACTG1
|
actin, gamma 1
|
|
chr17_-_77094375
|
1.621
|
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr20_-_62181231
|
1.619
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr16_+_66676833
|
1.600
|
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr7_-_103417198
|
1.594
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr15_+_78232160
|
1.593
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr14_-_90953883
|
1.592
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr9_-_115178969
|
1.590
|
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr7_-_150283794
|
1.586
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr7_-_154425510
|
1.577
|
NM_007349
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1
|
|
chr6_+_41622267
|
1.572
|
|
FOXP4
|
forkhead box P4
|
|
chr10_+_76256305
|
1.564
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr21_-_37260488
|
1.559
|
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr15_+_78232194
|
1.556
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr12_-_123914375
|
1.547
|
NM_001082959
NM_005505
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr16_+_29725665
|
1.526
|
|
|
|
|
chr5_-_134397740
|
1.515
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr20_+_31783426
|
1.511
|
NM_032819
|
ZNF341
|
zinc finger protein 341
|
|
chr21_+_43267760
|
1.509
|
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr20_+_30813851
|
1.499
|
NM_006892
NM_175848
NM_175849
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr2_-_32089309
|
1.494
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr17_+_26060617
|
1.488
|
|
|
|
|
chr2_-_32089226
|
1.487
|
|
MEMO1
|
mediator of cell motility 1
|
|
chr21_+_46473494
|
1.474
|
|
MCM3AP-AS1
|
MCM3AP antisense RNA 1 (non-protein coding)
|
|
chr19_-_14177980
|
1.470
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr19_-_53950458
|
1.457
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr11_+_550868
|
1.449
|
NM_001143993
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr2_+_112529270
|
1.444
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr1_+_945331
|
1.443
|
NM_198576
|
AGRN
|
agrin
|
|
chr19_-_50688310
|
1.440
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr12_+_56621711
|
1.440
|
NM_033276
|
XRCC6BP1
|
XRCC6 binding protein 1
|
|
chr12_-_123914219
|
1.437
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr1_+_172104115
|
1.434
|
NM_001122770
NM_032522
|
ZBTB37
|
zinc finger and BTB domain containing 37
|
|
chr22_-_22440054
|
1.431
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr7_+_151792052
|
1.426
|
|
LOC100128822
|
hypothetical LOC100128822
|
|
chr16_-_1369613
|
1.421
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr20_+_35407872
|
1.420
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr20_+_30410447
|
1.417
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr19_-_44034797
|
1.417
|
NM_001005335
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr6_+_41622111
|
1.411
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr15_-_20637731
|
1.406
|
NM_144599
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1
|
|
chr11_+_46358909
|
1.406
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr7_+_101860958
|
1.404
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr9_+_94860828
|
1.396
|
|
SUSD3
|
sushi domain containing 3
|
|
chr9_+_94860769
|
1.390
|
NM_145006
|
SUSD3
|
sushi domain containing 3
|
|
chr5_+_139008209
|
1.383
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr6_-_31977747
|
1.382
|
NM_181842
|
ZBTB12
|
zinc finger and BTB domain containing 12
|
|
chr7_-_156378382
|
1.376
|
|
LMBR1
|
limb region 1 homolog (mouse)
|
|
chr16_+_87047418
|
1.371
|
NM_153813
|
ZFPM1
|
zinc finger protein, multitype 1
|
|
chr6_+_169866714
|
1.371
|
|
LOC729088
|
hypothetical LOC729088
|
|
chr16_-_88295595
|
1.370
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr9_+_114288899
|
1.368
|
NM_133465
|
KIAA1958
|
KIAA1958
|
|
chr12_-_123914299
|
1.368
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr9_+_114289178
|
1.362
|
|
KIAA1958
|
KIAA1958
|
|
chr10_-_75241347
|
1.361
|
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2
|
|
chr16_+_88542639
|
1.360
|
NM_017702
NM_207514
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr7_+_137795719
|
1.359
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr20_-_3102169
|
1.357
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr3_+_185580554
|
1.353
|
NM_003741
|
CHRD
|
chordin
|
|
chr17_-_20886680
|
1.350
|
NM_015276
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr21_-_43369492
|
1.350
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr7_+_101861007
|
1.348
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr19_+_40223249
|
1.348
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr17_-_39632845
|
1.339
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr20_+_60906605
|
1.337
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr7_+_149696901
|
1.334
|
|
REPIN1
|
replication initiator 1
|
|
chr4_-_6253152
|
1.330
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr17_-_53946899
|
1.324
|
|
MTMR4
|
myotubularin related protein 4
|
|
chr16_-_30014993
|
1.320
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr6_+_149680747
|
1.314
|
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr2_+_218972721
|
1.312
|
NM_021198
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr6_+_7052985
|
1.312
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr22_-_49042065
|
1.311
|
NM_002969
|
MAPK12
|
mitogen-activated protein kinase 12
|
|
chr5_-_176671897
|
1.307
|
NM_001142935
NM_031300
|
MXD3
|
MAX dimerization protein 3
|
|
chr19_+_40900965
|
1.305
|
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr12_-_61614930
|
1.300
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr20_+_30814048
|
1.297
|
|
DNMT3B
|
DNA (cytosine-5-)-methyltransferase 3 beta
|
|
chr20_-_48980858
|
1.291
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr19_+_10404110
|
1.290
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_40224457
|
1.290
|
|
HPN
|
hepsin
|
|
chr12_+_49443903
|
1.285
|
NM_005171
|
ATF1
|
activating transcription factor 1
|
|
chr22_+_18081968
|
1.283
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr2_+_10101081
|
1.280
|
NM_003597
NM_001177716
|
KLF11
|
Kruppel-like factor 11
|
|
chr1_-_158334983
|
1.278
|
NM_052868
|
IGSF8
|
immunoglobulin superfamily, member 8
|
|
chr11_-_64268841
|
1.277
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr16_+_66676851
|
1.276
|
NM_004555
NM_173163
NM_173164
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr1_-_41480368
|
1.273
|
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chr16_+_2503915
|
1.272
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr20_+_61839931
|
1.271
|
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr2_+_233927880
|
1.270
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr2_-_61551350
|
1.270
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr18_+_11971421
|
1.268
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr7_-_150305933
|
1.264
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr14_+_74964261
|
1.263
|
NM_001135047
|
JDP2
|
Jun dimerization protein 2
|
|
chr6_+_11646472
|
1.257
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr19_+_59064471
|
1.257
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr11_-_72170533
|
1.257
|
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10
|
|
chr20_-_24986501
|
1.251
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr16_+_49139694
|
1.251
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr20_-_31737815
|
1.242
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr6_+_34312906
|
1.239
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_+_110328126
|
1.238
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr11_-_72141064
|
1.228
|
NM_001040118
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr2_+_233927892
|
1.226
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr11_-_16992524
|
1.226
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr1_+_90059992
|
1.220
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr14_+_101297887
|
1.219
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr19_-_2653662
|
1.203
|
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|