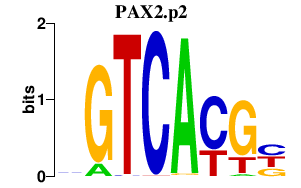
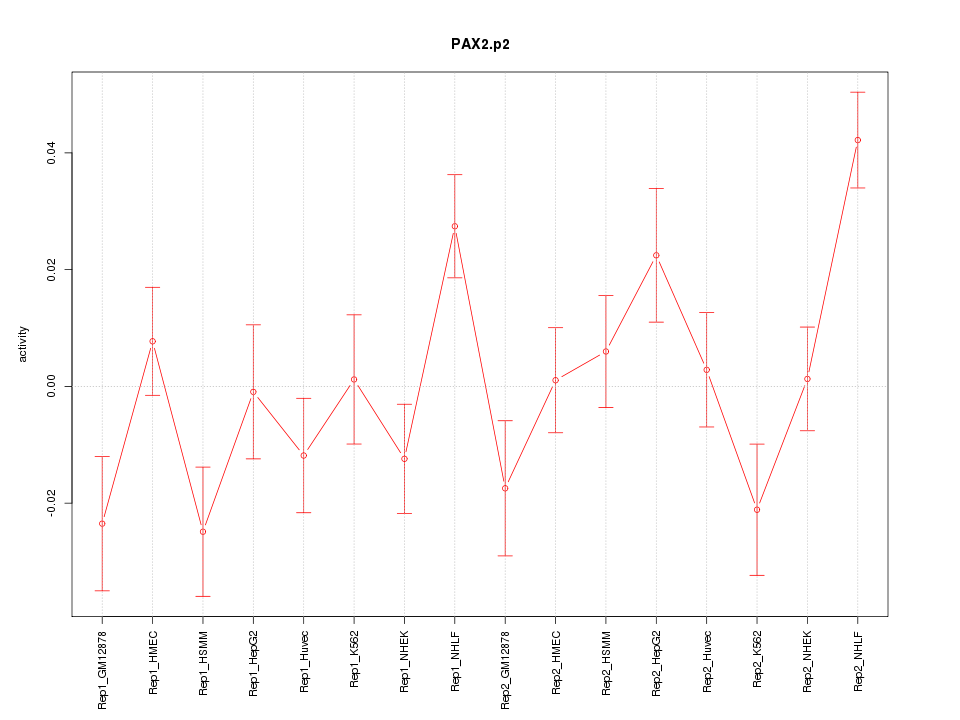
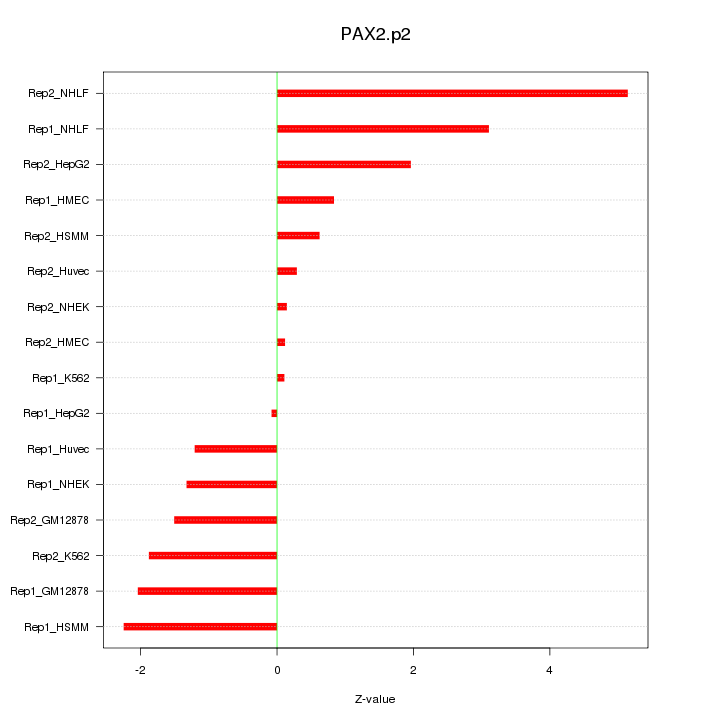
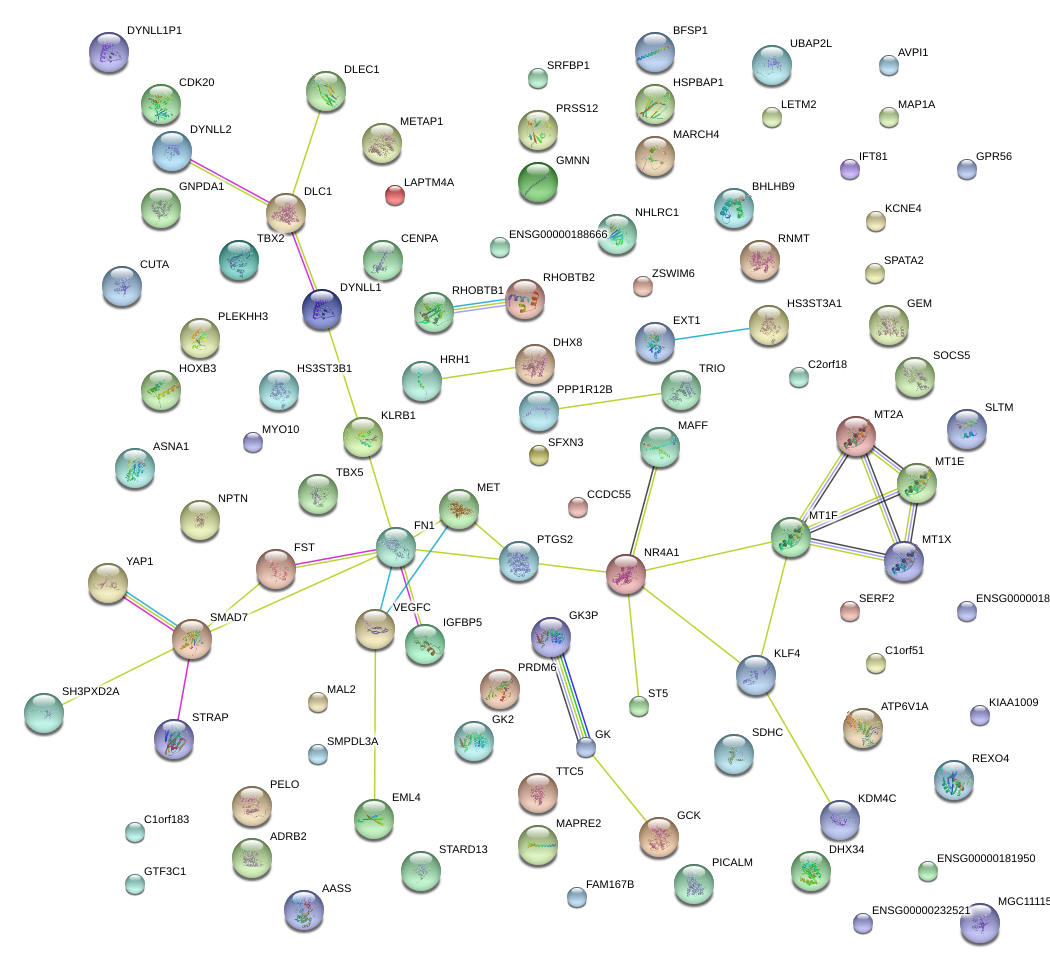
|
chr12_-_113330574
|
2.671
|
NM_000192
NM_080717
|
TBX5
|
T-box 5
|
|
chr17_-_44022595
|
1.867
|
|
HOXB3
|
homeobox B3
|
|
chr5_+_60663842
|
1.561
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr17_+_14145051
|
1.511
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr6_-_33493640
|
1.462
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr8_-_13178399
|
1.393
|
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_-_84994032
|
1.381
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr15_+_41597097
|
1.271
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr6_-_33493792
|
1.195
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr6_-_33493898
|
1.180
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr8_-_13178327
|
1.175
|
|
|
|
|
chr15_-_71712711
|
1.152
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr1_-_184916044
|
1.151
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr13_-_32757835
|
1.144
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_+_25467948
|
1.134
|
NM_032141
|
CCDC55
|
coiled-coil domain containing 55
|
|
chr6_-_33494042
|
1.110
|
NM_001014433
NM_001014837
NM_001014838
NM_001014840
NM_015921
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr8_-_119192985
|
1.108
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr12_+_15926554
|
1.101
|
NM_007178
|
STRAP
|
serine/threonine kinase receptor associated protein
|
|
chr8_-_95343716
|
1.089
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_-_16795336
|
1.075
|
|
MYO10
|
myosin X
|
|
chr8_-_95343696
|
1.060
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_-_216008689
|
1.020
|
|
FN1
|
fibronectin 1
|
|
chr12_+_15926681
|
1.006
|
|
STRAP
|
serine/threonine kinase receptor associated protein
|
|
chr10_+_102782151
|
0.974
|
|
SFXN3
|
sideroflexin 3
|
|
chr9_-_109291866
|
0.969
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr2_-_216009015
|
0.969
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr10_+_102782156
|
0.967
|
|
SFXN3
|
sideroflexin 3
|
|
chr22_+_36928972
|
0.958
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr9_-_89779180
|
0.956
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr12_+_50731457
|
0.928
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr11_+_101485946
|
0.923
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr3_+_114948550
|
0.910
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr2_+_46779827
|
0.904
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr3_-_123995216
|
0.900
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr20_-_17487480
|
0.899
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr3_+_11153778
|
0.898
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr18_+_30875297
|
0.880
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr5_+_148766632
|
0.877
|
|
LOC728264
|
hypothetical LOC728264
|
|
chr2_+_223625105
|
0.873
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr16_+_56219802
|
0.866
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr2_-_20115269
|
0.848
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr2_-_216944911
|
0.838
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr1_+_152459918
|
0.835
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr1_+_159550789
|
0.831
|
NM_001035511
NM_001035512
NM_001035513
NM_003001
|
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa
|
|
chr15_-_71712605
|
0.829
|
|
NPTN
|
neuroplastin
|
|
chr8_+_103889733
|
0.827
|
|
FLJ45248
|
FLJ45248 protein
|
|
chr5_+_52812165
|
0.826
|
|
FST
|
follistatin
|
|
chr16_-_27468717
|
0.821
|
|
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa
|
|
chr11_-_85457469
|
0.816
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_+_6747919
|
0.815
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr5_+_14493905
|
0.814
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr12_+_15926743
|
0.812
|
|
STRAP
|
serine/threonine kinase receptor associated protein
|
|
chr5_+_52812236
|
0.799
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr5_+_52131686
|
0.798
|
|
PELO
|
pelota homolog (Drosophila)
|
|
chr4_-_177950658
|
0.795
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr8_+_120289735
|
0.787
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr10_-_99436889
|
0.785
|
NM_021732
|
AVPI1
|
arginine vasopressin-induced 1
|
|
chr11_-_8695974
|
0.785
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_+_42249896
|
0.779
|
NM_001145076
NM_019063
|
EML4
|
echinoderm microtubule associated protein like 4
|
|
chr20_-_47965393
|
0.772
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr4_-_119493322
|
0.762
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr1_+_200584421
|
0.750
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr9_-_89779413
|
0.749
|
NM_001039803
NM_001170639
NM_001170640
NM_012119
NM_178432
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr12_+_109046522
|
0.748
|
NM_001143779
NM_014055
NM_031473
|
IFT81
|
intraflagellar transport 81 homolog (Chlamydomonas)
|
|
chr17_+_56831951
|
0.747
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr7_+_116099648
|
0.728
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr5_-_141372721
|
0.722
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chrX_+_30581523
|
0.717
|
|
GK
|
glycerol kinase
|
|
chr3_+_114948600
|
0.715
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr5_+_148186348
|
0.714
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr1_-_112083387
|
0.711
|
NM_019099
|
C1orf183
|
chromosome 1 open reading frame 183
|
|
chr19_+_12709281
|
0.709
|
NM_004317
|
ASNA1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial)
|
|
chr3_+_114948591
|
0.701
|
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr16_+_55199978
|
0.698
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr1_+_148521852
|
0.690
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr15_+_41871768
|
0.688
|
NM_001018108
|
SERF2
|
small EDRK-rich factor 2
|
|
chr18_-_44729700
|
0.685
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_26840626
|
0.685
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr10_-_105427898
|
0.681
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr6_+_123152044
|
0.681
|
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chrX_+_101862297
|
0.670
|
NM_001142524
NM_001142525
NM_001142526
NM_001142527
NM_001142528
NM_030639
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9
|
|
chr1_+_32485404
|
0.668
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr10_-_62373828
|
0.668
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr2_-_217267742
|
0.665
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr6_-_18230829
|
0.659
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr17_-_38081967
|
0.653
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr8_-_13416554
|
0.652
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr5_+_122452739
|
0.651
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr8_+_38363150
|
0.651
|
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr7_-_121571472
|
0.651
|
NM_005763
|
AASS
|
aminoadipate-semialdehyde synthase
|
|
chr15_+_41871809
|
0.647
|
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 read-through transcript
small EDRK-rich factor 2
|
|
chr11_+_101291134
|
0.643
|
|
KIAA1377
|
KIAA1377
|
|
chr1_-_59020875
|
0.642
|
|
JUN
|
jun proto-oncogene
|
|
chr18_-_53621277
|
0.642
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr9_+_116389918
|
0.637
|
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr8_-_38505331
|
0.630
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr6_+_52393763
|
0.629
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr1_+_152460381
|
0.625
|
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr12_-_113328271
|
0.622
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr4_+_77446906
|
0.618
|
|
STBD1
|
starch binding domain 1
|
|
chr5_-_150118747
|
0.615
|
NM_001135643
NM_016221
|
DCTN4
|
dynactin 4 (p62)
|
|
chr7_+_28415495
|
0.611
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr2_-_227372574
|
0.609
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr1_+_152644668
|
0.607
|
|
IL6R
|
interleukin 6 receptor
|
|
chr3_-_188946168
|
0.606
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr19_-_52709975
|
0.606
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chrX_+_30581377
|
0.605
|
NM_000167
NM_001128127
NM_203391
|
GK
|
glycerol kinase
|
|
chr3_-_36961539
|
0.595
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr11_+_101290930
|
0.590
|
NM_020802
|
KIAA1377
|
KIAA1377
|
|
chr20_-_47965474
|
0.590
|
NM_006038
|
SPATA2
|
spermatogenesis associated 2
|
|
chr3_-_188945921
|
0.590
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr4_+_120029426
|
0.581
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr12_-_79855580
|
0.580
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr9_+_119506446
|
0.579
|
|
TLR4
|
toll-like receptor 4
|
|
chr5_-_150118274
|
0.577
|
NM_001135644
|
DCTN4
|
dynactin 4 (p62)
|
|
chr1_-_37792414
|
0.567
|
NM_024700
|
SNIP1
|
Smad nuclear interacting protein 1
|
|
chr3_-_188492163
|
0.566
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr15_-_41572660
|
0.563
|
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr22_+_34025872
|
0.562
|
|
TOM1
|
target of myb1 (chicken)
|
|
chr20_-_47965443
|
0.560
|
|
SPATA2
|
spermatogenesis associated 2
|
|
chr5_-_64813459
|
0.559
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr16_+_27468942
|
0.558
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chr17_-_39556483
|
0.556
|
|
HDAC5
|
histone deacetylase 5
|
|
chr5_+_150807331
|
0.551
|
NM_078483
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr14_-_80757327
|
0.549
|
NM_201595
|
GTF2A1
|
general transcription factor IIA, 1, 19/37kDa
|
|
chr9_-_98421894
|
0.549
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr17_+_72795567
|
0.548
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr20_+_2744947
|
0.547
|
NM_001167670
|
LOC100288797
|
hypothetical protein LOC100288797
|
|
chr5_+_150807397
|
0.545
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr17_-_63965156
|
0.543
|
NM_017983
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr17_-_39556535
|
0.542
|
NM_001015053
NM_005474
|
HDAC5
|
histone deacetylase 5
|
|
chr4_+_81337663
|
0.542
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr11_-_8788765
|
0.542
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr3_+_67131416
|
0.538
|
NM_032505
|
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8
|
|
chr16_+_28893525
|
0.537
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr11_+_117982422
|
0.534
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr5_-_90645974
|
0.525
|
|
LOC100505994
|
hypothetical LOC100505994
|
|
chr9_+_116389814
|
0.525
|
NM_004888
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr8_-_27528215
|
0.524
|
NM_001831
|
CLU
|
clusterin
|
|
chr2_+_87536174
|
0.522
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr11_+_61479182
|
0.514
|
|
BEST1
|
bestrophin 1
|
|
chr1_-_20685314
|
0.513
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr14_+_19993181
|
0.512
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr6_+_148705421
|
0.512
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr8_-_27528105
|
0.511
|
|
CLU
|
clusterin
|
|
chr9_-_13269562
|
0.511
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr4_+_81406765
|
0.511
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr12_+_28234657
|
0.511
|
|
|
|
|
chr20_-_2592777
|
0.510
|
NM_006899
NM_174855
NM_174856
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr15_-_99609197
|
0.509
|
|
CHSY1
|
chondroitin sulfate synthase 1
|
|
chr15_-_41572558
|
0.508
|
NM_001141979
NM_001141980
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr7_-_158304062
|
0.504
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr12_+_27568311
|
0.502
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr11_-_85457753
|
0.499
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr1_+_36462583
|
0.496
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr12_+_14818572
|
0.495
|
|
H2AFJ
|
H2A histone family, member J
|
|
chr11_-_85457736
|
0.495
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_62902279
|
0.494
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr1_-_152459697
|
0.494
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr20_+_1254033
|
0.493
|
|
LOC100507495
|
hypothetical LOC100507495
|
|
chr11_-_62213577
|
0.493
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr16_+_2023279
|
0.493
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr16_+_65868913
|
0.492
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chr13_-_49597776
|
0.489
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr11_-_82582328
|
0.489
|
|
DKFZp547G183
|
hypothetical LOC55525
|
|
chr16_-_69277355
|
0.485
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr1_+_205137410
|
0.484
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr5_+_96064248
|
0.483
|
NM_173060
|
CAST
|
calpastatin
|
|
chr5_+_150807380
|
0.481
|
|
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1
|
|
chr16_-_65742323
|
0.472
|
NM_033309
|
B3GNT9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9
|
|
chr22_+_40107878
|
0.472
|
NM_003216
|
TEF
|
thyrotrophic embryonic factor
|
|
chr22_+_39583001
|
0.471
|
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr6_+_112515366
|
0.470
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr4_-_39199527
|
0.468
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr10_+_128583984
|
0.467
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr8_+_109525050
|
0.467
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr12_-_121276513
|
0.466
|
|
DIABLO
|
diablo homolog (Drosophila)
|
|
chr6_+_116939500
|
0.465
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr5_+_80292313
|
0.464
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr6_+_32229771
|
0.463
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr13_+_47776026
|
0.462
|
|
RB1
|
retinoblastoma 1
|
|
chr17_+_30594192
|
0.458
|
NM_144975
|
SLFN5
|
schlafen family member 5
|
|
chr8_+_109525010
|
0.458
|
NM_014673
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr1_-_220952276
|
0.458
|
NM_022831
|
AIDA
|
axin interactor, dorsalization associated
|
|
chr8_-_145090892
|
0.457
|
NM_201381
|
PLEC
|
plectin
|
|
chr6_-_112515329
|
0.456
|
NM_016262
|
TUBE1
|
tubulin, epsilon 1
|
|
chr13_-_20533703
|
0.456
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr15_-_68781662
|
0.454
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_+_185536376
|
0.454
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr1_+_36462625
|
0.453
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_-_18175731
|
0.453
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr19_+_60688349
|
0.453
|
NM_020378
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative)
|
|
chr10_+_102662675
|
0.453
|
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr11_+_64948860
|
0.452
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr1_-_68470585
|
0.451
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr3_+_30622905
|
0.450
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr5_+_80292213
|
0.449
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr12_-_106011437
|
0.449
|
|
CRY1
|
cryptochrome 1 (photolyase-like)
|
|
chr13_-_20533629
|
0.449
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_-_45914618
|
0.448
|
NM_024876
|
ADCK4
|
aarF domain containing kinase 4
|