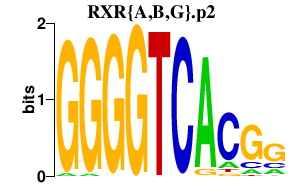
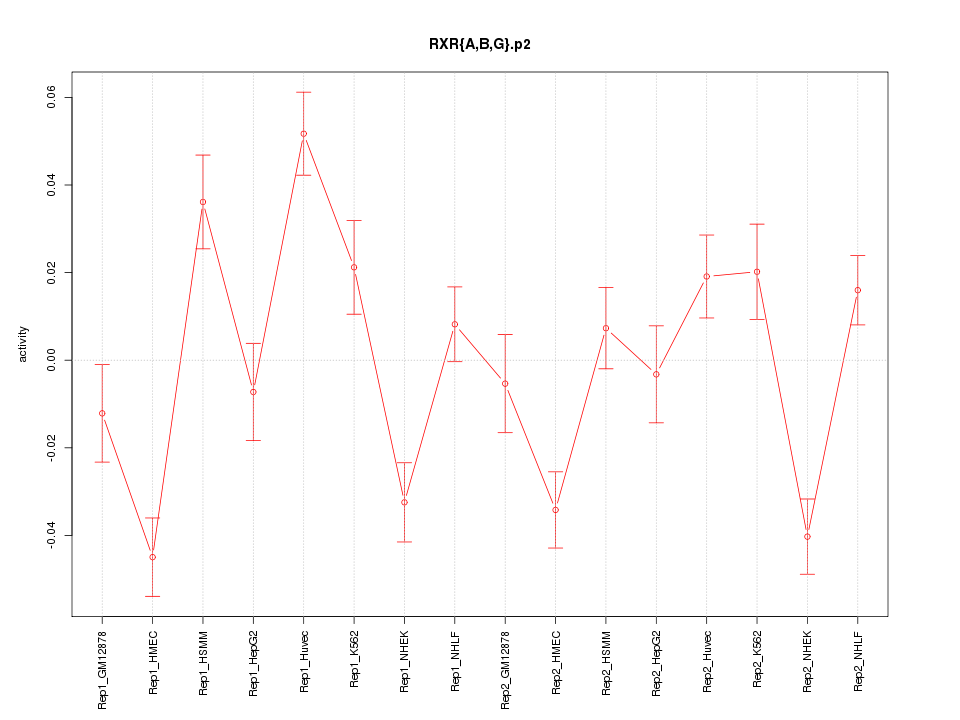
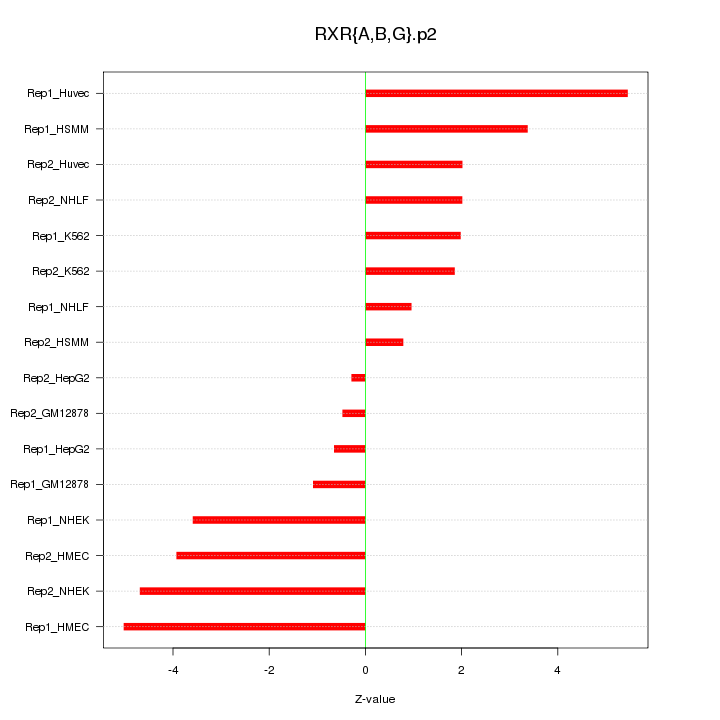
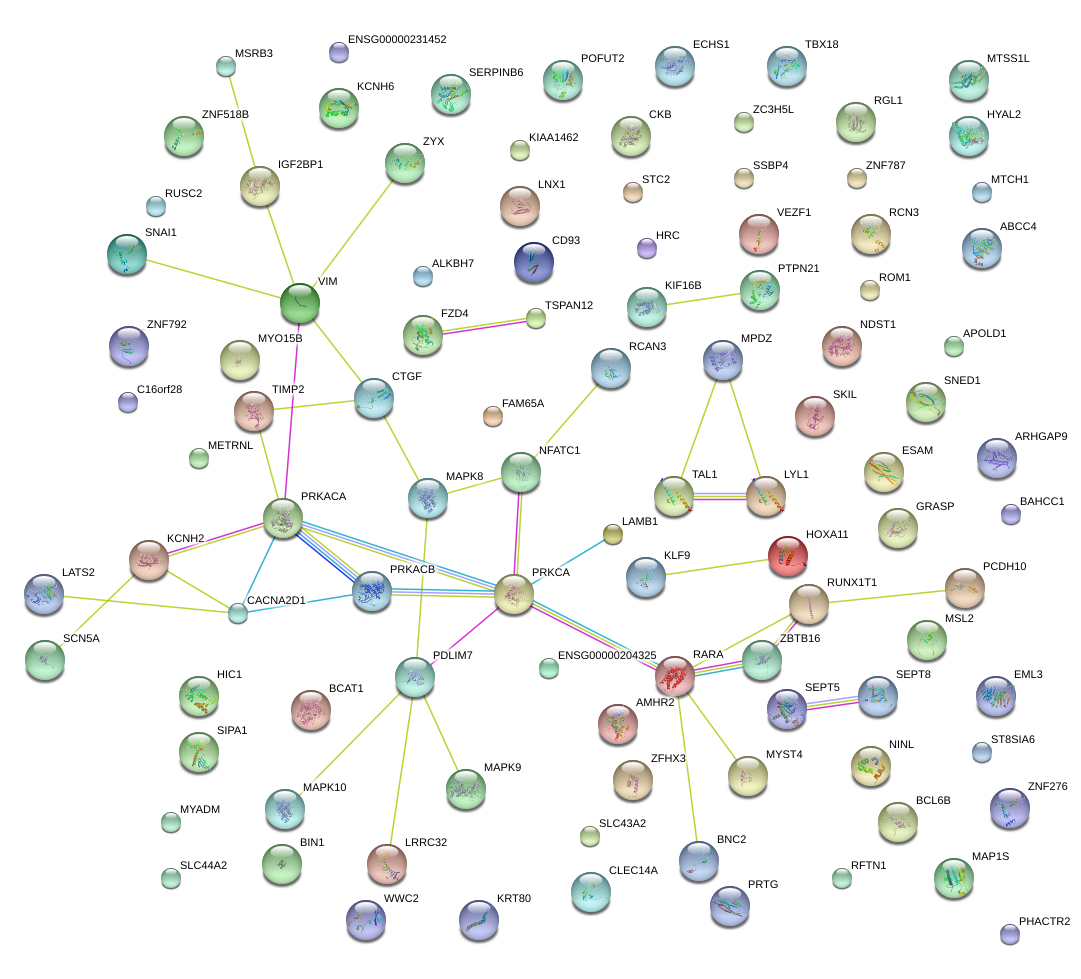
|
chr17_+_44429748
|
6.269
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr19_+_18391211
|
2.939
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr21_-_45532176
|
2.934
|
NM_015227
NM_133635
|
POFUT2
|
protein O-fucosyltransferase 2
|
|
chr19_-_14089560
|
2.559
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr5_+_149845503
|
2.549
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr6_-_2916786
|
2.548
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr19_+_18391423
|
2.473
|
|
|
|
|
chr6_-_132314004
|
2.443
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr6_-_2916280
|
2.321
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr19_-_14089362
|
2.277
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr11_+_65162114
|
2.089
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr17_+_6867090
|
2.016
|
NM_181844
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr18_+_75261231
|
1.955
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr3_+_129690735
|
1.949
|
|
|
|
|
chr12_+_50686990
|
1.917
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr15_-_53822468
|
1.893
|
NM_173814
|
PRTG
|
protogenin
|
|
chr5_-_172687798
|
1.886
|
|
STC2
|
stanniocalcin 2
|
|
chr1_-_47468029
|
1.875
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr3_-_16529996
|
1.823
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr14_-_37795001
|
1.818
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr22_+_18081968
|
1.816
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr20_+_48032918
|
1.792
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr17_+_35751796
|
1.779
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr17_+_6867138
|
1.776
|
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr10_+_17310475
|
1.734
|
|
VIM
|
vimentin
|
|
chr5_-_172688024
|
1.730
|
|
STC2
|
stanniocalcin 2
|
|
chr16_-_71650034
|
1.723
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr16_-_69277355
|
1.706
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr21_+_45649719
|
1.689
|
|
|
|
|
chr10_-_135036805
|
1.678
|
NM_004092
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial
|
|
chr20_-_23014828
|
1.642
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr14_-_37795290
|
1.641
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr12_-_24946495
|
1.497
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr19_-_40146707
|
1.495
|
|
ZNF792
|
zinc finger protein 792
|
|
chr11_-_86343859
|
1.474
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr16_+_66129481
|
1.472
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr22_+_18085891
|
1.471
|
|
SEPT5
|
septin 5
|
|
chr11_-_86343737
|
1.464
|
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr17_+_76988134
|
1.459
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr3_-_38666122
|
1.458
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr4_-_10068055
|
1.453
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chr6_-_85529031
|
1.444
|
|
TBX18
|
T-box 18
|
|
chr8_-_93184629
|
1.428
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_50335113
|
1.422
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr11_+_113436438
|
1.420
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr4_+_184257337
|
1.387
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr22_+_18085990
|
1.385
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr11_-_62136778
|
1.372
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr19_-_40146792
|
1.363
|
NM_175872
|
ZNF792
|
zinc finger protein 792
|
|
chr10_+_76256305
|
1.361
|
NM_012330
|
MYST4
|
MYST histone acetyltransferase (monocytic leukemia) 4
|
|
chr11_-_124137260
|
1.321
|
NM_138961
|
ESAM
|
endothelial cell adhesion molecule
|
|
chr2_+_241586927
|
1.281
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr4_+_134292630
|
1.280
|
|
PCDH10
|
protocadherin 10
|
|
chr12_+_12829807
|
1.279
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr7_-_107430566
|
1.268
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr19_+_17691302
|
1.254
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr17_+_1905104
|
1.245
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr4_+_134293171
|
1.239
|
|
PCDH10
|
protocadherin 10
|
|
chr6_-_37061902
|
1.203
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr5_-_132141368
|
1.198
|
NM_001098813
|
SEPT8
|
septin 8
|
|
chr17_-_74433048
|
1.171
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr6_+_144040751
|
1.165
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr14_-_103058568
|
1.157
|
|
CKB
|
creatine kinase, brain
|
|
chr19_+_59064471
|
1.139
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_-_13269562
|
1.133
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chr20_-_25514137
|
1.123
|
NM_025176
|
NINL
|
ninein-like
|
|
chr19_+_59064795
|
1.119
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr3_+_171558215
|
1.117
|
|
SKIL
|
SKI-like oncogene
|
|
chr5_-_176857158
|
1.112
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr10_-_30388439
|
1.106
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr19_+_6323443
|
1.103
|
NM_032306
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli)
|
|
chr11_-_76059435
|
1.100
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr17_+_78630793
|
1.096
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr6_-_37061625
|
1.094
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr21_+_45532377
|
1.086
|
|
LOC642852
|
hypothetical LOC642852
|
|
chr5_-_176857207
|
1.081
|
NM_005451
NM_203352
NM_213636
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr19_-_13074973
|
1.070
|
NM_005583
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr17_-_1478859
|
1.069
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr19_-_54350457
|
1.067
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr3_+_171558188
|
1.060
|
|
SKIL
|
SKI-like oncogene
|
|
chr19_-_55046718
|
1.058
|
|
LOC100506033
|
hypothetical LOC100506033
|
|
chr9_-_16860719
|
1.056
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr12_+_63958689
|
1.055
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr7_-_27191287
|
1.050
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr19_+_54723044
|
1.043
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr10_-_17536181
|
1.028
|
NM_001004470
|
ST8SIA6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6
|
|
chr3_+_171558143
|
1.024
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr6_-_37061897
|
1.014
|
|
MTCH1
|
mitochondrial carrier homolog 1 (C. elegans)
|
|
chr13_-_20533703
|
1.013
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr13_-_94751573
|
1.009
|
NM_001105515
NM_005845
|
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4
|
|
chr11_+_62136788
|
1.005
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr19_+_10574055
|
1.004
|
NM_001145056
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr7_-_120285245
|
0.996
|
|
TSPAN12
|
tetraspanin 12
|
|
chr10_+_49184658
|
0.993
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
chr16_-_1369613
|
0.989
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr16_+_88315452
|
0.987
|
NM_001113525
|
ZNF276
|
zinc finger protein 276
|
|
chr19_-_13074669
|
0.982
|
|
LYL1
|
lymphoblastic leukemia derived sequence 1
|
|
chr19_-_61324412
|
0.978
|
|
ZNF787
|
zinc finger protein 787
|
|
chr7_-_81910956
|
0.978
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr2_-_127580975
|
0.976
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr10_+_17310263
|
0.976
|
NM_003380
|
VIM
|
vimentin
|
|
chr17_-_53420473
|
0.974
|
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr9_-_72218098
|
0.972
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr12_+_52105131
|
0.970
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr2_+_113709553
|
0.968
|
|
LOC654433
|
hypothetical LOC654433
|
|
chr5_-_132140786
|
0.966
|
NM_001098811
NM_001098812
NM_015146
|
SEPT8
|
septin 8
|
|
chr7_-_150283794
|
0.962
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr9_+_35480018
|
0.960
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr17_+_71097147
|
0.958
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr7_+_142788521
|
0.957
|
|
ZYX
|
zyxin
|
|
chr3_-_137397344
|
0.951
|
NM_018133
|
MSL2
|
male-specific lethal 2 homolog (Drosophila)
|
|
chr14_-_88090641
|
0.949
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr1_+_182040837
|
0.944
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr17_-_53420595
|
0.944
|
NM_007146
|
VEZF1
|
vascular endothelial zinc finger 1
|
|
chr12_-_50871953
|
0.942
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr9_+_35479989
|
0.935
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr9_+_35782405
|
0.926
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr7_-_81910740
|
0.926
|
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr11_-_61415429
|
0.925
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr17_-_1478293
|
0.925
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr15_-_46725087
|
0.925
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr19_-_55046117
|
0.924
|
|
LOC100506033
|
hypothetical LOC100506033
|
|
chr19_+_17691283
|
0.917
|
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr13_-_20533629
|
0.913
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr12_+_50587468
|
0.911
|
NM_000020
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr7_+_27191534
|
0.911
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr9_-_16860664
|
0.906
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_59131210
|
0.905
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr5_-_92942680
|
0.904
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr2_-_144991556
|
0.899
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_1341599
|
0.898
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr19_+_16860807
|
0.895
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr7_+_78920846
|
0.890
|
|
LOC100505881
|
hypothetical LOC100505881
|
|
chr19_-_50840446
|
0.890
|
NM_001193268
|
EML2
|
echinoderm microtubule associated protein like 2
|
|
chr19_-_61324456
|
0.886
|
NM_001002836
|
ZNF787
|
zinc finger protein 787
|
|
chr12_+_63958949
|
0.886
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr20_+_30409649
|
0.884
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr5_+_72504778
|
0.884
|
NM_153217
|
TMEM174
|
transmembrane protein 174
|
|
chr10_+_94439658
|
0.883
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr1_-_152185764
|
0.878
|
NM_014856
|
DENND4B
|
DENN/MADD domain containing 4B
|
|
chr17_+_78286687
|
0.877
|
NM_022158
|
FN3K
|
fructosamine 3 kinase
|
|
chr2_-_74583850
|
0.875
|
NM_001009812
|
LBX2
|
ladybird homeobox 2
|
|
chr16_-_87244896
|
0.874
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr16_+_85169615
|
0.872
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr5_-_172594691
|
0.870
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr12_+_105873669
|
0.869
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr1_-_36562341
|
0.868
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr7_-_75206212
|
0.866
|
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chrX_-_153397566
|
0.864
|
NM_001171132
NM_001171133
NM_001171134
NM_021806
|
FAM3A
|
family with sequence similarity 3, member A
|
|
chr10_+_97794139
|
0.864
|
|
|
|
|
chr1_+_26896519
|
0.863
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr10_+_80569464
|
0.850
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr17_-_74432671
|
0.840
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr11_+_67563093
|
0.831
|
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr9_-_101621964
|
0.830
|
|
|
|
|
chr18_+_12648191
|
0.827
|
|
|
|
|
chr19_-_11169457
|
0.820
|
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr19_-_40938251
|
0.819
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr2_+_74583762
|
0.819
|
|
LOC151534
|
hypothetical LOC151534
|
|
chr11_-_129377929
|
0.819
|
NM_020228
NM_199437
|
PRDM10
|
PR domain containing 10
|
|
chr7_-_2561638
|
0.815
|
|
BAAT1
|
BRCA1-associated ATM activator 1
|
|
chr2_+_56265260
|
0.812
|
|
|
|
|
chr6_+_117109042
|
0.810
|
NM_002269
|
KPNA5
|
karyopherin alpha 5 (importin alpha 6)
|
|
chr19_+_55046088
|
0.809
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr3_-_16530132
|
0.803
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr10_-_103444899
|
0.799
|
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chrX_+_55495262
|
0.795
|
NM_014061
|
MAGEH1
|
melanoma antigen family H, 1
|
|
chr10_+_11099860
|
0.794
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_27155197
|
0.791
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr12_+_130979696
|
0.791
|
NM_001002020
|
PUS1
|
pseudouridylate synthase 1
|
|
chr10_-_44816265
|
0.789
|
NM_001039380
|
C10orf25
|
chromosome 10 open reading frame 25
|
|
chr17_-_7106987
|
0.787
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr17_-_74432800
|
0.785
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr1_+_12149662
|
0.784
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr5_+_170221441
|
0.783
|
|
RANBP17
|
RAN binding protein 17
|
|
chr12_-_95317928
|
0.782
|
|
CDK17
|
cyclin-dependent kinase 17
|
|
chr11_+_65049552
|
0.782
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr17_-_37681837
|
0.778
|
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr7_-_75206152
|
0.771
|
|
HIP1
|
huntingtin interacting protein 1
|
|
chr19_+_55046229
|
0.770
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr7_-_106088565
|
0.768
|
NM_175884
|
FLJ36031
|
hypothetical protein FLJ36031
|
|
chr10_+_44816278
|
0.768
|
NM_006963
|
ZNF22
|
zinc finger protein 22 (KOX 15)
|
|
chr6_+_43846372
|
0.766
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr2_-_192767494
|
0.766
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr3_-_121652183
|
0.759
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr19_-_10537646
|
0.755
|
|
KRI1
|
KRI1 homolog (S. cerevisiae)
|
|
chr19_-_2378581
|
0.754
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr7_-_27190882
|
0.754
|
|
HOXA11
|
homeobox A11
|
|
chr17_+_63799228
|
0.752
|
|
ARSG
|
arylsulfatase G
|
|
chr17_-_37681917
|
0.749
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr2_-_175337386
|
0.749
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr6_-_2916980
|
0.746
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr2_+_190916440
|
0.746
|
NM_001128928
NM_002194
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr19_+_6312462
|
0.743
|
NM_006012
|
CLPP
|
ClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit homolog (E. coli)
|
|
chr14_+_95575319
|
0.739
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr11_+_111313067
|
0.738
|
NM_001037954
|
DIXDC1
|
DIX domain containing 1
|
|
chr7_+_27190688
|
0.737
|
|
|
|
|
chr5_-_180169720
|
0.737
|
NM_001114617
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr7_-_16427471
|
0.732
|
NM_001101417
NM_001101426
|
ISPD
|
isoprenoid synthase domain containing
|
|
chr7_-_78920806
|
0.727
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|