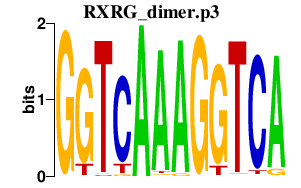
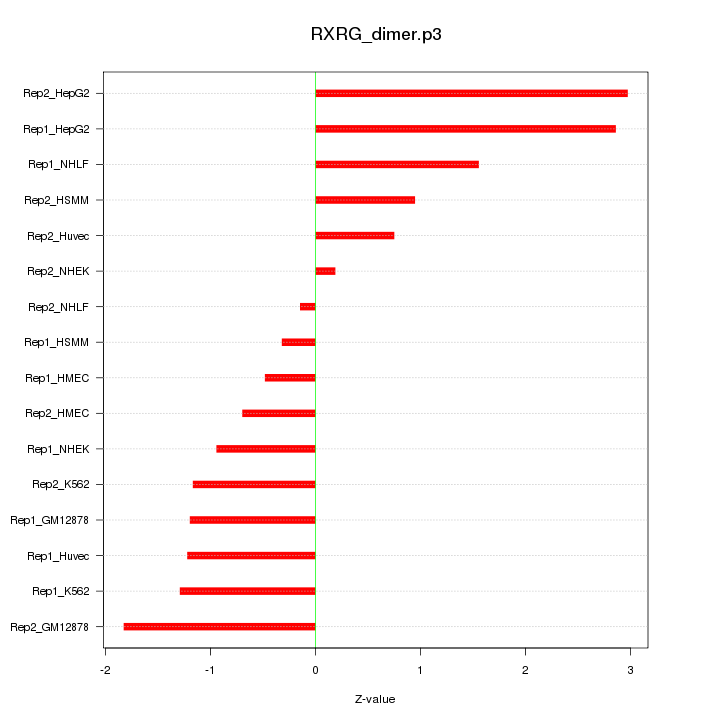
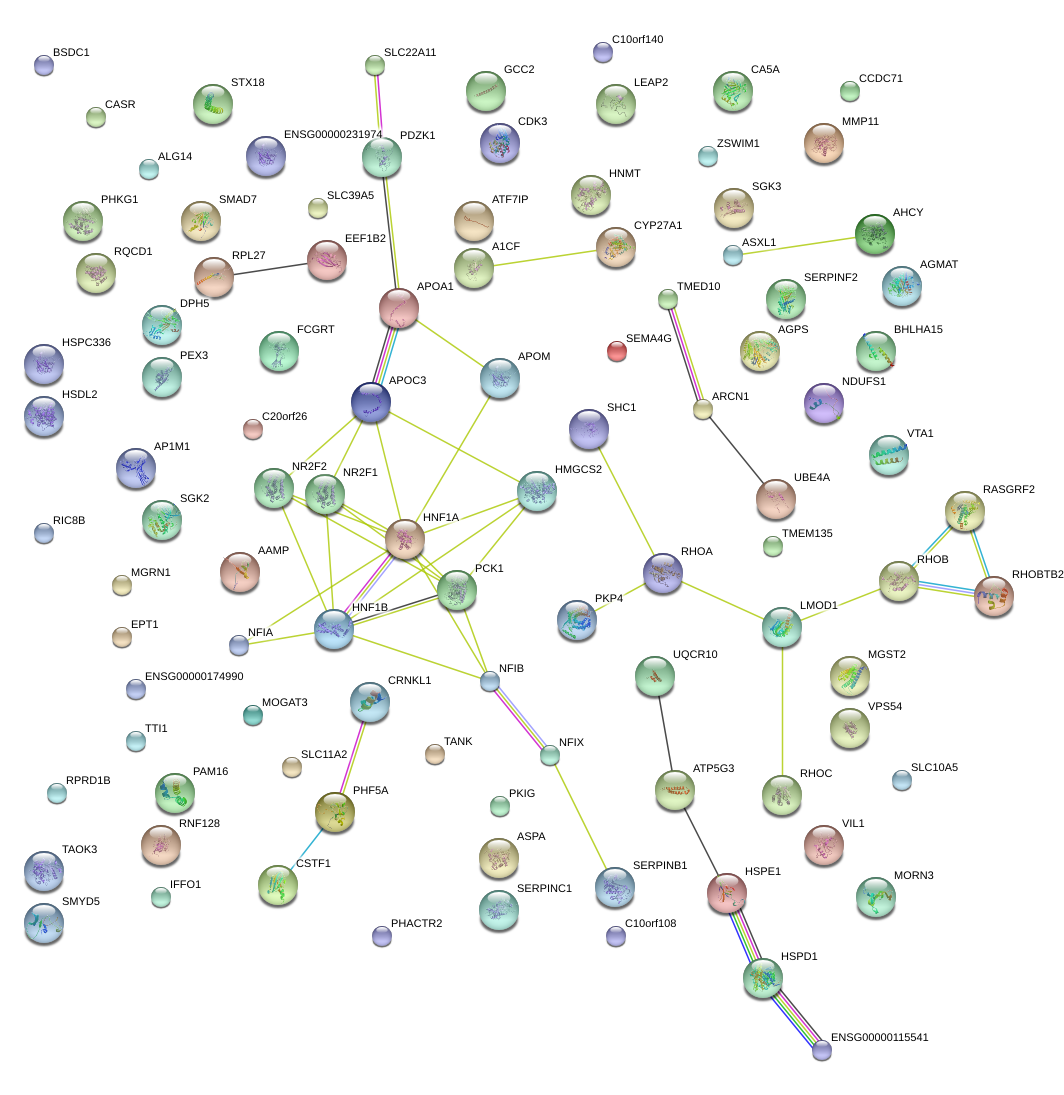
|
chr11_+_116205830
|
5.022
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr11_-_116213511
|
3.155
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr16_-_86527608
|
2.788
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr2_+_138438277
|
2.747
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr16_+_4654710
|
2.728
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chr7_-_100630912
|
2.660
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr5_+_92944680
|
2.628
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr20_+_55569542
|
2.548
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr12_+_54910767
|
2.524
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr2_+_73294852
|
2.242
|
NM_006062
|
SMYD5
|
SMYD family member 5
|
|
chr10_+_102722275
|
2.205
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr2_+_218992053
|
2.138
|
NM_007127
|
VIL1
|
villin 1
|
|
chr6_+_31731667
|
2.137
|
|
APOM
|
apolipoprotein M
|
|
chr6_+_31731647
|
2.136
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr20_+_41621079
|
2.119
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr20_+_41621119
|
2.018
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr5_-_42847716
|
2.000
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr12_+_119900677
|
1.950
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr10_-_52315390
|
1.793
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr2_+_161725172
|
1.744
|
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr5_+_80292313
|
1.613
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr10_-_21854616
|
1.608
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr2_-_206732348
|
1.607
|
NM_005006
|
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase)
|
|
chr2_+_206732575
|
1.597
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr6_+_30702586
|
1.548
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr2_+_206732562
|
1.526
|
NM_001037663
NM_001959
NM_021121
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr12_+_119900931
|
1.489
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr17_-_33178987
|
1.446
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr10_-_52315436
|
1.410
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr18_-_44730706
|
1.405
|
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_206732859
|
1.376
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr1_+_144439069
|
1.376
|
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr6_+_144040751
|
1.375
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr11_+_86426518
|
1.373
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr2_+_219354692
|
1.298
|
NM_000784
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr1_+_61320213
|
1.290
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr17_-_33179118
|
1.211
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr2_+_177965734
|
1.184
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr1_-_172153087
|
1.178
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr20_-_19981005
|
1.154
|
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr17_+_38403935
|
1.072
|
NM_000988
|
RPL27
|
ribosomal protein L27
|
|
chr20_+_19981157
|
1.052
|
NM_015585
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr18_-_44731078
|
1.022
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr19_+_54708302
|
0.993
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr19_+_54708312
|
0.935
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr12_-_6535481
|
0.889
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr1_-_172153024
|
0.886
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr17_+_38403991
|
0.885
|
|
RPL27
|
ribosomal protein L27
|
|
chr2_+_177965617
|
0.877
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr4_+_140806536
|
0.861
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr2_-_64099935
|
0.834
|
|
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae)
|
|
chr19_+_16169664
|
0.821
|
NM_001130524
NM_032493
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr20_-_32363268
|
0.817
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr1_-_15784048
|
0.812
|
NM_024758
|
AGMAT
|
agmatine ureohydrolase (agmatinase)
|
|
chr6_-_2785682
|
0.808
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr5_+_132237253
|
0.806
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr2_-_177965626
|
0.793
|
|
LOC100130691
|
hypothetical LOC100130691
|
|
chr12_+_14429499
|
0.770
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr17_+_3324096
|
0.760
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr15_+_94674849
|
0.744
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr10_+_685887
|
0.740
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr11_+_117735529
|
0.728
|
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr2_+_108432097
|
0.723
|
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr2_+_198072965
|
0.722
|
NM_002157
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr11_+_117735502
|
0.721
|
NM_004788
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr17_+_71508581
|
0.710
|
NM_001258
|
CDK3
|
cyclin-dependent kinase 3
|
|
chr2_+_108432008
|
0.698
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr14_-_20642214
|
0.681
|
|
|
|
|
chr22_+_22445035
|
0.678
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr11_+_117735530
|
0.665
|
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr11_+_64079665
|
0.659
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr6_+_142510075
|
0.652
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr12_-_117295095
|
0.648
|
NM_016281
|
TAOK3
|
TAO kinase 3
|
|
chr2_+_198073358
|
0.626
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr2_+_159021653
|
0.624
|
|
PKP4
|
plakophilin 4
|
|
chr2_-_198072805
|
0.609
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr12_-_49706364
|
0.594
|
NM_000617
NM_001174126
NM_001174127
NM_001174128
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr6_+_142510094
|
0.571
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr2_-_198072767
|
0.553
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr6_+_142510034
|
0.551
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr20_+_36095263
|
0.547
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr3_-_49424322
|
0.545
|
|
RHOA
|
ras homolog gene family, member A
|
|
chrX_+_105823723
|
0.527
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr2_-_218843109
|
0.513
|
NM_001087
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr2_+_198073257
|
0.503
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr11_+_117735558
|
0.501
|
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr6_+_142510108
|
0.500
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr17_+_1592893
|
0.484
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr1_-_32632601
|
0.480
|
NM_001143888
NM_001143889
NM_001143890
NM_018045
|
BSDC1
|
BSD domain containing 1
|
|
chr8_-_82769761
|
0.478
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr7_+_97679501
|
0.475
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr2_-_198073242
|
0.472
|
NM_199440
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr3_-_49424380
|
0.467
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr20_+_42593849
|
0.462
|
NM_007066
NM_181804
NM_181805
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr22_+_28493351
|
0.461
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr1_-_95311031
|
0.460
|
NM_144988
|
ALG14
|
asparagine-linked glycosylation 14 homolog (S. cerevisiae)
|
|
chr20_+_54400979
|
0.453
|
NM_001324
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr9_+_114182216
|
0.446
|
|
HSDL2
|
hydroxysteroid dehydrogenase like 2
|
|
chr11_+_117948301
|
0.442
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr3_-_49178738
|
0.439
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr1_-_120112989
|
0.438
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr3_-_49178762
|
0.432
|
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr9_+_114182009
|
0.431
|
NM_001195822
NM_032303
|
HSDL2
|
hydroxysteroid dehydrogenase like 2
|
|
chr2_-_218843088
|
0.430
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr2_-_198072868
|
0.420
|
NM_002156
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr3_-_49424397
|
0.419
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424361
|
0.410
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr1_-_153213454
|
0.408
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr2_-_218843039
|
0.405
|
|
AAMP
|
angio-associated, migratory cell protein
|
|
chr3_-_49178773
|
0.403
|
NM_022903
|
CCDC71
|
coiled-coil domain containing 71
|
|
chr20_+_36095552
|
0.402
|
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr2_+_26422495
|
0.400
|
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr3_-_49424413
|
0.400
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr3_-_49424427
|
0.397
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr20_+_36095621
|
0.395
|
|
|
|
|
chr14_-_74713062
|
0.393
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr12_+_105692466
|
0.386
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr16_-_4341266
|
0.384
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr8_+_22909305
|
0.379
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr14_-_74713086
|
0.374
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr20_-_36095236
|
0.373
|
NM_014657
|
TTI1
|
Tel2 interacting protein 1 homolog (S. pombe)
|
|
chr12_-_49706190
|
0.372
|
NM_001174129
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr12_-_117294933
|
0.369
|
|
TAOK3
|
TAO kinase 3
|
|
chr1_-_101263867
|
0.367
|
NM_001077394
NM_001077395
NM_015958
|
DPH5
|
DPH5 homolog (S. cerevisiae)
|
|
chr19_+_54708247
|
0.353
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr20_+_43943254
|
0.352
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr20_+_30409649
|
0.350
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr11_+_117948350
|
0.341
|
|
ARCN1
|
archain 1
|
|
chr2_-_175754366
|
0.332
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr1_-_153213425
|
0.329
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr4_-_4594591
|
0.329
|
|
STX18
|
syntaxin 18
|
|
chr2_+_219141761
|
0.327
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr17_+_1592879
|
0.322
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr4_+_140806371
|
0.320
|
NM_002413
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr3_-_49424404
|
0.318
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr14_-_74713088
|
0.318
|
NM_006827
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr16_+_31105579
|
0.314
|
|
|
|
|
chr6_+_143813472
|
0.313
|
NM_003630
|
PEX3
|
peroxisomal biogenesis factor 3
|
|
chr19_+_54708389
|
0.310
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr22_-_40194602
|
0.305
|
NM_032758
|
PHF5A
|
PHD finger protein 5A
|
|
chr3_-_114947330
|
0.296
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr14_-_74713039
|
0.295
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr14_-_94693359
|
0.293
|
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr16_-_3433490
|
0.291
|
NM_152457
|
ZNF597
|
zinc finger protein 597
|
|
chr10_-_62431203
|
0.288
|
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr14_-_20636337
|
0.286
|
|
ZNF219
|
zinc finger protein 219
|
|
chr9_+_103335983
|
0.277
|
|
RNF20
|
ring finger protein 20
|
|
chr1_+_78126787
|
0.273
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr4_-_84150885
|
0.268
|
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr22_+_48927610
|
0.262
|
NM_001164106
|
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse)
|
|
chr3_-_49424506
|
0.258
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr1_-_158579653
|
0.256
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr1_-_158579680
|
0.249
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr17_+_7474128
|
0.246
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr4_-_4594620
|
0.244
|
|
STX18
|
syntaxin 18
|
|
chr2_-_24160683
|
0.243
|
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr3_-_49424529
|
0.237
|
NM_001664
|
RHOA
|
ras homolog gene family, member A
|
|
chr2_+_119841611
|
0.234
|
NM_001079863
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein)
|
|
chr2_+_27967116
|
0.232
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr4_-_102487356
|
0.230
|
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme
|
|
chr10_+_75174125
|
0.229
|
NM_004922
NM_198597
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chr1_+_1351370
|
0.228
|
NM_001146685
|
TMEM88B
|
transmembrane protein 88B
|
|
chr14_-_74713012
|
0.226
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr13_+_20612709
|
0.221
|
|
SAP18
|
Sin3A-associated protein, 18kDa
|
|
chr9_+_103335964
|
0.215
|
|
RNF20
|
ring finger protein 20
|
|
chr1_-_158579583
|
0.209
|
|
|
|
|
chrX_-_64171317
|
0.205
|
NM_001178032
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr2_+_27967102
|
0.204
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr6_+_32034767
|
0.196
|
|
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae)
|
|
chrX_+_106030049
|
0.195
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr2_+_27966985
|
0.194
|
NM_199193
NM_199194
NM_004899
NM_199191
NM_199192
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr20_-_54400628
|
0.193
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr14_+_94693744
|
0.191
|
|
FLJ45244
|
hypothetical locus FLJ45244
|
|
chr18_+_27852785
|
0.186
|
|
RNF125
|
ring finger protein 125
|
|
chr10_+_75174151
|
0.182
|
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chr17_+_7472000
|
0.179
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr1_+_45046740
|
0.178
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chrX_+_106057984
|
0.177
|
|
CLDN2
|
claudin 2
|
|
chr14_-_20636568
|
0.173
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr17_-_40344322
|
0.169
|
|
|
|
|
chr12_-_49708279
|
0.169
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr1_-_153213483
|
0.162
|
NM_001130041
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr12_-_6949877
|
0.161
|
|
PHB2
|
prohibitin 2
|
|
chr4_-_84150997
|
0.160
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr4_+_4594619
|
0.157
|
|
LOC100507266
|
hypothetical LOC100507266
|
|
chrX_+_100241453
|
0.156
|
NM_006733
|
CENPI
|
centromere protein I
|
|
chr12_-_93991235
|
0.153
|
NM_001032287
NM_001127362
NM_003297
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1
|
|
chr17_+_35532994
|
0.153
|
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr6_-_32034607
|
0.151
|
NM_002904
|
RDBP
|
RD RNA binding protein
|
|
chr9_+_103335941
|
0.150
|
NM_019592
|
RNF20
|
ring finger protein 20
|
|
chr1_+_158579685
|
0.149
|
NM_015331
|
NCSTN
|
nicastrin
|
|
chr6_-_146326918
|
0.149
|
NM_001042683
NM_173082
|
SHPRH
|
SNF2 histone linker PHD RING helicase
|
|
chr9_-_115178161
|
0.147
|
NM_031219
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr7_+_288195
|
0.146
|
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr22_+_40195058
|
0.143
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr2_-_27966715
|
0.141
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr2_-_159021201
|
0.140
|
NM_001171637
NM_138803
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr2_+_27967159
|
0.140
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr3_-_114947681
|
0.139
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr11_+_65140207
|
0.138
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|