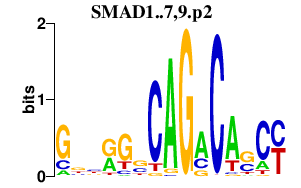
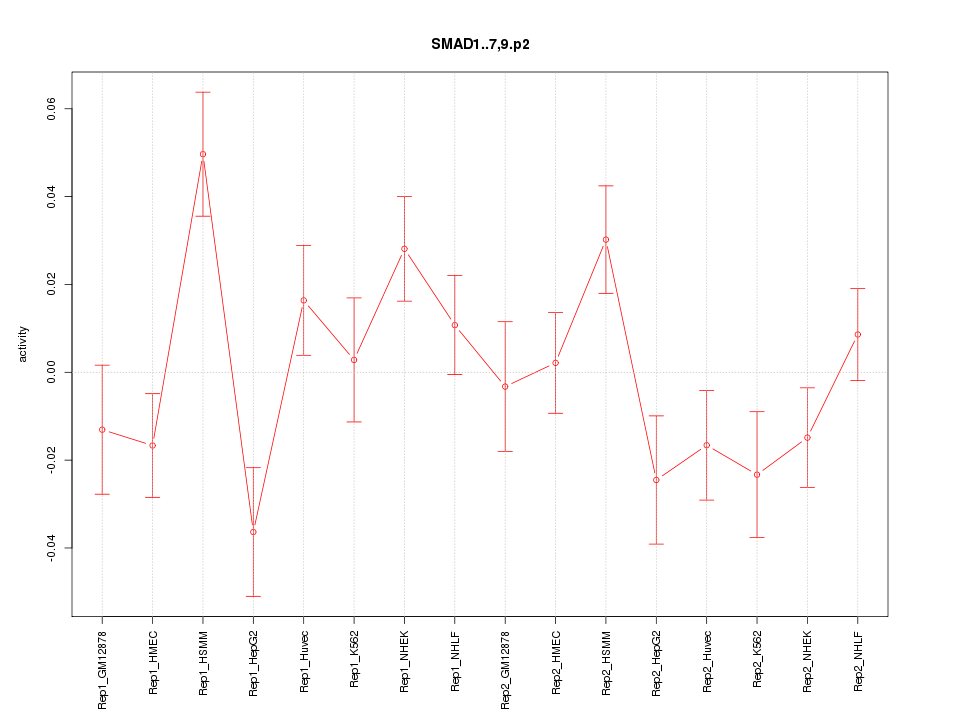
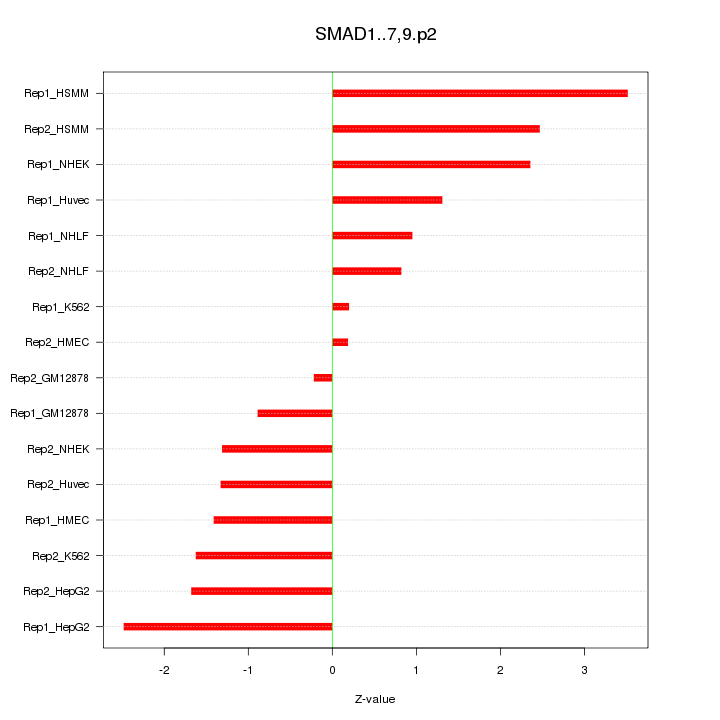
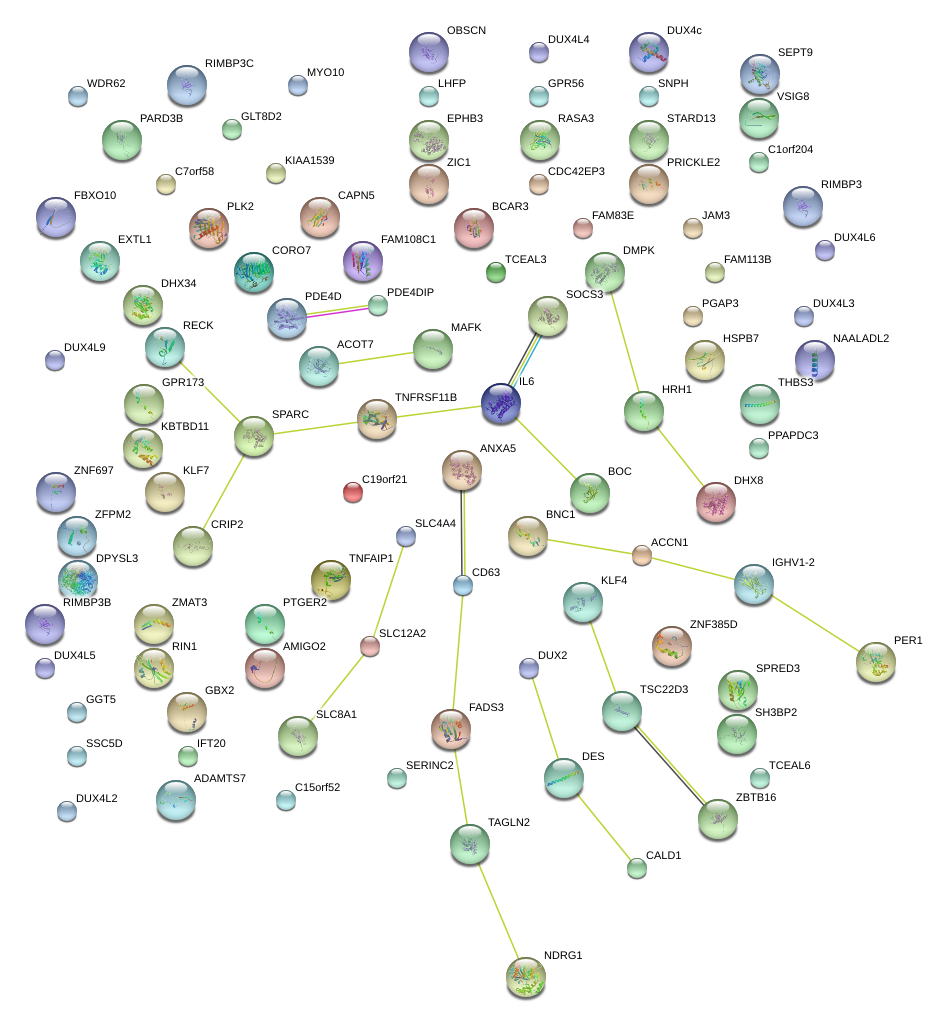
|
chr22_-_22970850
|
3.465
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr15_-_81744469
|
2.562
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr13_-_32822759
|
2.202
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr5_-_151046638
|
2.174
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr1_+_15145001
|
2.144
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr19_+_60691733
|
2.119
|
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr5_-_151046680
|
2.063
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr16_+_56219802
|
2.047
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr10_+_22765996
|
1.869
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr11_-_65860450
|
1.775
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr12_-_45759691
|
1.743
|
NM_001143668
NM_181847
|
AMIGO2
|
adhesion molecule with Ig-like domain 2
|
|
chrX_+_53095230
|
1.499
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr9_+_133154896
|
1.439
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr3_+_175641340
|
1.399
|
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr1_-_6368324
|
1.389
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr14_+_51850754
|
1.382
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr19_+_60691671
|
1.377
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr9_+_36026900
|
1.250
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr1_-_16217871
|
1.235
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr19_-_50977586
|
1.233
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr22_-_18841785
|
1.196
|
NM_015672
|
RIMBP3
|
RIMS binding protein 3
|
|
chr1_-_143750988
|
1.179
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr8_+_106400322
|
1.167
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr12_+_45759634
|
1.163
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr3_+_11171213
|
1.151
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr5_-_58370693
|
1.144
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_73867748
|
1.140
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr2_+_219991342
|
1.134
|
NM_001927
|
DES
|
desmin
|
|
chr7_+_120416103
|
1.125
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr4_+_191226073
|
1.091
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr4_+_191229360
|
1.083
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr9_-_109291575
|
1.066
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr10_+_135343649
|
1.056
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr4_+_191235959
|
1.051
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chr7_+_1543644
|
1.042
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr17_-_23686566
|
1.037
|
NM_174887
|
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas)
|
|
chr1_-_93919789
|
1.013
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr5_+_127447306
|
1.010
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr19_+_702108
|
0.989
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chrX_-_106846253
|
0.978
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_31659246
|
0.949
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr3_+_114414064
|
0.932
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr12_-_102967828
|
0.929
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr15_-_38420414
|
0.905
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr17_+_23686674
|
0.903
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr9_-_35101592
|
0.896
|
|
KIAA1539
|
KIAA1539
|
|
chr14_+_105012106
|
0.895
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr19_-_50977592
|
0.890
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr19_-_50977485
|
0.889
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr2_+_205118801
|
0.884
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr22_+_20067662
|
0.883
|
NM_001128633
|
RIMBP3C
|
RIMS binding protein 3C
|
|
chr9_-_90456824
|
0.879
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr15_-_76890626
|
0.877
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr1_-_197173159
|
0.877
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr1_-_158161877
|
0.876
|
|
TAGLN2
|
transgelin 2
|
|
chr4_-_122837590
|
0.874
|
NM_001154
|
ANXA5
|
annexin A5
|
|
chr1_-_158091760
|
0.872
|
NM_001134233
|
C1orf204
|
chromosome 1 open reading frame 204
|
|
chr12_-_102968044
|
0.867
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr19_-_53808505
|
0.847
|
NM_017708
|
FAM83E
|
family with sequence similarity 83, member E
|
|
chr4_+_191239247
|
0.845
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191232653
|
0.841
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr1_+_26220852
|
0.839
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr2_-_40532651
|
0.828
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr7_+_1546535
|
0.826
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr15_+_78774664
|
0.808
|
NM_021214
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr3_+_148610516
|
0.805
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr13_-_113916066
|
0.801
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr10_+_135330357
|
0.799
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr11_-_61415429
|
0.798
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr2_-_236741352
|
0.789
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr1_-_153444304
|
0.788
|
NM_007112
|
THBS3
|
thrombospondin 3
|
|
chrX_+_102749489
|
0.786
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr4_+_72271218
|
0.786
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_7996409
|
0.782
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr11_+_76455574
|
0.777
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr4_+_191242540
|
0.774
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr3_-_64186151
|
0.769
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr1_-_119991912
|
0.769
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr20_+_1194959
|
0.762
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr1_+_204049053
|
0.760
|
|
|
|
|
chr17_-_45629401
|
0.755
|
|
|
|
|
chr5_-_146813343
|
0.755
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr8_+_1936549
|
0.751
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr4_+_2783764
|
0.749
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr13_+_66702683
|
0.744
|
|
|
|
|
chr13_-_113916115
|
0.743
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr11_+_113436438
|
0.732
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr3_-_180272291
|
0.730
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr3_+_185762233
|
0.730
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chrX_-_106846684
|
0.729
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_-_127447197
|
0.726
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr2_-_207738858
|
0.721
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr3_-_21767819
|
0.718
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr2_+_56265260
|
0.718
|
|
|
|
|
chr2_-_37752272
|
0.718
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr7_+_22733290
|
0.716
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr9_-_37566248
|
0.712
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr12_+_56406296
|
0.710
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr5_-_16989290
|
0.709
|
|
MYO10
|
myosin X
|
|
chr8_-_134378630
|
0.702
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr16_-_4406608
|
0.692
|
|
CORO7
|
coronin 7
|
|
chr13_-_39075304
|
0.692
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr16_-_4406596
|
0.689
|
|
CORO7
|
coronin 7
|
|
chr8_-_120033240
|
0.687
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr12_-_54409710
|
0.685
|
|
CD63
|
CD63 molecule
|
|
chr5_-_57791544
|
0.685
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr7_+_134114957
|
0.681
|
|
CALD1
|
caldesmon 1
|
|
chr19_+_43572679
|
0.681
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr11_+_133444029
|
0.678
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr4_-_1192749
|
0.678
|
NM_001199021
|
LOC100130872
SPON2
|
hypothetical LOC100130872
spondin 2, extracellular matrix protein
|
|
chr2_+_30307900
|
0.674
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr3_-_197103848
|
0.671
|
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr1_-_154913712
|
0.668
|
NM_006617
|
NES
|
nestin
|
|
chr12_-_51759434
|
0.668
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr10_-_73203202
|
0.666
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr22_+_29333069
|
0.666
|
NM_000355
NM_001184726
|
TCN2
|
transcobalamin II
|
|
chr22_+_44828338
|
0.665
|
|
|
|
|
chr1_-_32036858
|
0.662
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr12_-_51759400
|
0.655
|
|
SPRYD3
|
SPRY domain containing 3
|
|
chr15_+_78775008
|
0.652
|
|
FAM108C1
|
family with sequence similarity 108, member C1
|
|
chr22_-_20235749
|
0.650
|
NM_001128633
|
RIMBP3C
|
RIMS binding protein 3C
|
|
chr9_+_91409746
|
0.650
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chrX_+_48915127
|
0.645
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr11_-_1974398
|
0.645
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_+_26220895
|
0.644
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr11_-_109672401
|
0.644
|
NM_002906
|
RDX
|
radixin
|
|
chr1_+_78126787
|
0.643
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr5_+_126654354
|
0.642
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr15_-_68971665
|
0.637
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr1_+_182622850
|
0.628
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr21_-_35343065
|
0.624
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_158161902
|
0.624
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chrX_+_2680092
|
0.623
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chr17_-_1342700
|
0.618
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr16_+_3295433
|
0.616
|
NM_153028
|
ZNF75A
|
zinc finger protein 75a
|
|
chr1_-_150276091
|
0.616
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr7_+_134114701
|
0.609
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr16_-_4406571
|
0.606
|
|
CORO7-PAM16
CORO7
|
CORO7-PAM16 read-through transcript
coronin 7
|
|
chr8_+_22493928
|
0.604
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr1_-_32036802
|
0.602
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr11_-_124175493
|
0.601
|
NM_024631
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr3_+_11153778
|
0.598
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr4_+_109072120
|
0.596
|
NM_183075
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1
|
|
chr1_-_31618405
|
0.595
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr9_-_37455384
|
0.591
|
NM_014872
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr1_+_221955876
|
0.589
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr17_-_1335763
|
0.588
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr16_+_88467518
|
0.587
|
|
TCF25
|
transcription factor 25 (basic helix-loop-helix)
|
|
chr10_-_43464157
|
0.583
|
NM_001005368
|
ZNF32
|
zinc finger protein 32
|
|
chr12_-_51759438
|
0.582
|
NM_032840
|
SPRYD3
|
SPRY domain containing 3
|
|
chr16_-_63713382
|
0.581
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr1_+_15935395
|
0.580
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr15_-_58477407
|
0.579
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr15_+_97010219
|
0.579
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr10_+_135333667
|
0.579
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_-_60573552
|
0.579
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr11_-_60475558
|
0.579
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr8_+_1759502
|
0.579
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr8_-_145085616
|
0.578
|
NM_201384
|
PLEC
|
plectin
|
|
chr1_-_150276064
|
0.577
|
|
S100A11
|
S100 calcium binding protein A11
|
|
chr16_-_30992323
|
0.574
|
NM_001172668
|
ZNF668
|
zinc finger protein 668
|
|
chr1_-_150276114
|
0.573
|
NM_005620
|
S100A11
|
S100 calcium binding protein A11
|
|
chr17_-_18102767
|
0.560
|
NM_002018
|
FLII
|
flightless I homolog (Drosophila)
|
|
chr3_-_16530132
|
0.559
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr7_-_44195405
|
0.558
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr7_+_32963486
|
0.558
|
NM_007270
|
FKBP9
|
FK506 binding protein 9, 63 kDa
|
|
chr2_-_161058550
|
0.557
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_-_158161898
|
0.556
|
|
TAGLN2
|
transgelin 2
|
|
chr10_-_33663777
|
0.554
|
NM_001024628
NM_001024629
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr17_-_7238546
|
0.554
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr7_+_120415958
|
0.553
|
NM_024913
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr19_-_58092624
|
0.552
|
|
ZNF320
|
zinc finger protein 320
|
|
chr10_+_112247614
|
0.547
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr4_+_151218862
|
0.546
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr10_-_128066935
|
0.544
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr2_+_205118710
|
0.544
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chrX_+_100361588
|
0.544
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr3_-_180272229
|
0.543
|
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr11_-_1973768
|
0.542
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr17_-_34607257
|
0.541
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr11_-_1975583
|
0.540
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_+_112337199
|
0.537
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr16_-_4406632
|
0.536
|
NM_024535
|
CORO7
|
coronin 7
|
|
chr17_-_18102594
|
0.536
|
|
FLII
|
flightless I homolog (Drosophila)
|
|
chr9_-_122516399
|
0.535
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr15_-_81037554
|
0.533
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr7_-_76667045
|
0.532
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr10_-_125841840
|
0.531
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr1_+_199426575
|
0.530
|
NM_001164586
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
|
chr1_+_182622751
|
0.527
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr8_-_105670300
|
0.522
|
|
LRP12
|
low density lipoprotein receptor-related protein 12
|
|
chr15_-_58706934
|
0.522
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr11_-_124175467
|
0.519
|
|
C11orf61
|
chromosome 11 open reading frame 61
|
|
chr1_-_158161924
|
0.518
|
|
TAGLN2
|
transgelin 2
|
|
chr14_-_67211081
|
0.517
|
NM_006370
|
VTI1B
|
vesicle transport through interaction with t-SNAREs homolog 1B (yeast)
|
|
chr9_-_138772506
|
0.515
|
NM_178469
|
LCN8
|
lipocalin 8
|
|
chrX_-_77801480
|
0.514
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr13_+_23451919
|
0.514
|
|
SPATA13
|
spermatogenesis associated 13
|
|
chr19_-_50975603
|
0.513
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr1_-_10779291
|
0.512
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|