|
chr18_-_26935883
|
1.129
|
NM_004949
NM_024422
|
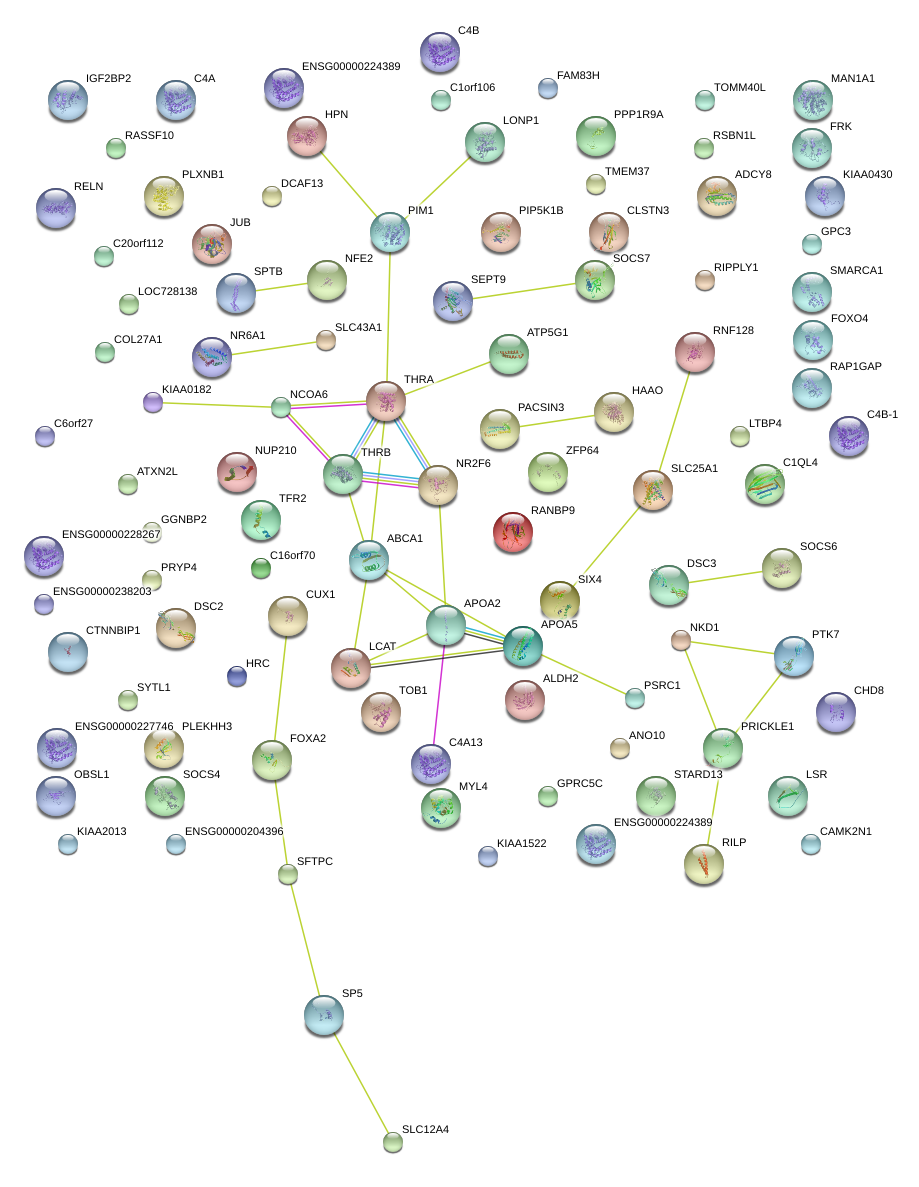
DSC2
|
desmocollin 2
|
|
chr12_-_52980914
|
1.038
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr17_+_42641712
|
0.910
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr2_-_220144254
|
0.868
|
|
OBSL1
|
obscurin-like 1
|
|
chr8_-_144887876
|
0.800
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr19_+_40431678
|
0.750
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr20_-_42457717
|
0.697
|
|
|
|
|
chr7_-_103417198
|
0.693
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr6_-_116488458
|
0.684
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr9_+_70509925
|
0.683
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr1_+_32992131
|
0.677
|
|
|
|
|
chr1_-_11909042
|
0.674
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr16_+_65701284
|
0.670
|
NM_025187
|
C16orf70
|
chromosome 16 open reading frame 70
|
|
chr3_-_24511456
|
0.668
|
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr12_-_52980984
|
0.655
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chrX_+_105856531
|
0.638
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr16_-_66535457
|
0.635
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr16_+_65701428
|
0.629
|
|
|
|
|
chr3_-_24511180
|
0.614
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr2_+_119905812
|
0.607
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr20_-_30636535
|
0.600
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr3_-_13436802
|
0.568
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chrX_-_128485121
|
0.568
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr6_-_138231006
|
0.544
|
|
|
|
|
chr1_-_21868380
|
0.535
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_-_54350457
|
0.524
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr17_+_42641426
|
0.508
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr19_+_40223249
|
0.498
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr6_-_13819463
|
0.485
|
|
RANBP9
|
RAN binding protein 9
|
|
chr19_+_45799060
|
0.477
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr1_-_20684858
|
0.473
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr17_+_7682355
|
0.460
|
|
|
|
|
chr1_-_9892852
|
0.459
|
NM_001012329
NM_020248
|
CTNNBIP1
|
catenin, beta interacting protein 1
|
|
chr17_+_33761234
|
0.444
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr7_+_94374862
|
0.442
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr1_+_159462456
|
0.437
|
NM_032174
|
TOMM40L
|
translocase of outer mitochondrial membrane 40 homolog (yeast)-like
|
|
chr1_+_199130571
|
0.433
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr20_-_22513050
|
0.426
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr17_+_72795567
|
0.422
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr16_+_84202515
|
0.422
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr3_-_13436725
|
0.421
|
|
NUP210
|
nucleoporin 210kDa
|
|
chr14_-_20975241
|
0.409
|
NM_020920
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr1_-_109627290
|
0.408
|
NM_001005290
NM_001032291
NM_032636
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chrX_-_132947290
|
0.406
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr4_-_187884721
|
0.405
|
|
|
|
|
chr19_+_40431618
|
0.403
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chrX_+_70232362
|
0.401
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr1_-_159459999
|
0.400
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr3_-_48445719
|
0.393
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr3_-_187025438
|
0.392
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr6_-_31853027
|
0.390
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr14_-_22516271
|
0.387
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr7_+_77163665
|
0.385
|
NM_198467
|
RSBN1L
|
round spermatid basic protein 1-like
|
|
chr1_+_32979930
|
0.383
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chr6_+_32090516
|
0.381
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr6_+_37245860
|
0.379
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chrX_-_132947141
|
0.378
|
|
GPC3
|
glypican 3
|
|
chr20_-_32877029
|
0.375
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr19_+_40431359
|
0.371
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr14_-_60260780
|
0.369
|
|
SIX4
|
SIX homeobox 4
|
|
chr1_+_27541086
|
0.369
|
|
SYTL1
|
synaptotagmin-like 1
|
|
chr9_-_106730157
|
0.361
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_-_109627266
|
0.355
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr1_-_109627206
|
0.353
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr19_-_17217022
|
0.353
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr17_-_1499849
|
0.351
|
NM_031430
|
RILP
|
Rab interacting lysosomal protein
|
|
chr7_+_101247601
|
0.345
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chrX_-_128485062
|
0.340
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr17_+_31975611
|
0.339
|
|
GGNBP2
|
gametogenetin binding protein 2
|
|
chr16_-_66535923
|
0.339
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr13_-_33148899
|
0.338
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_-_48017223
|
0.338
|
NM_001008223
|
C1QL4
|
complement component 1, q subcomponent-like 4
|
|
chr12_+_7174230
|
0.337
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr17_-_38081967
|
0.335
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr16_+_28741861
|
0.332
|
|
ATXN2L
|
ataxin 2-like
|
|
chr17_+_69939806
|
0.331
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr2_+_171280944
|
0.331
|
|
SP5
|
Sp5 transcription factor
|
|
chr11_+_12987455
|
0.330
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr11_-_116168316
|
0.327
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr6_+_43151922
|
0.326
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr17_+_44325127
|
0.325
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr3_-_187025501
|
0.324
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr7_-_100077072
|
0.322
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr6_-_119712596
|
0.318
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr14_-_64416307
|
0.317
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr2_-_42873179
|
0.317
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr9_-_126573349
|
0.317
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr11_-_47164514
|
0.316
|
NM_001184974
NM_016223
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr20_-_50241581
|
0.315
|
|
ZFP64
|
zinc finger protein 64 homolog (mouse)
|
|
chr16_+_49139694
|
0.311
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr17_-_46298671
|
0.307
|
|
TOB1
|
transducer of ERBB2, 1
|
|
chr11_-_57039721
|
0.306
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chrX_-_106033202
|
0.305
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr16_-_15644465
|
0.303
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr22_-_17546234
|
0.302
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr9_+_115957537
|
0.301
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr2_+_219780192
|
0.299
|
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr12_+_56621581
|
0.299
|
|
XRCC6BP1
|
XRCC6 binding protein 1
|
|
chr16_-_4262696
|
0.296
|
|
TFAP4
|
transcription factor AP-4 (activating enhancer binding protein 4)
|
|
chr17_-_36469869
|
0.296
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr17_-_40858775
|
0.295
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr1_+_166172679
|
0.295
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr8_+_76482661
|
0.294
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr6_+_30959839
|
0.294
|
NM_001954
|
DDR1
|
discoidin domain receptor tyrosine kinase 1
|
|
chr17_-_24302597
|
0.290
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr19_-_6671585
|
0.286
|
NM_000064
|
C3
|
complement component 3
|
|
chr14_+_34521854
|
0.286
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr2_+_219779737
|
0.286
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr22_+_38720881
|
0.281
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr17_+_35147697
|
0.281
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr6_+_150112537
|
0.280
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr7_-_77163475
|
0.280
|
|
|
|
|
chr14_+_34521945
|
0.276
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr12_+_52665196
|
0.274
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr11_+_117812282
|
0.274
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr1_-_42573210
|
0.270
|
NM_001198851
|
FOXJ3
|
forkhead box J3
|
|
chr6_+_150112511
|
0.268
|
NM_005389
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr17_-_24302450
|
0.267
|
|
PHF12
|
PHD finger protein 12
|
|
chr4_-_22126437
|
0.265
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr2_-_220144427
|
0.264
|
|
OBSL1
|
obscurin-like 1
|
|
chr1_+_227473458
|
0.263
|
NM_004578
|
RAB4A
|
RAB4A, member RAS oncogene family
|
|
chr14_-_22974707
|
0.263
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr16_+_2954217
|
0.262
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr3_-_95175282
|
0.262
|
|
PROS1
|
protein S (alpha)
|
|
chr16_+_29819647
|
0.262
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr3_-_159306214
|
0.262
|
|
|
|
|
chr19_-_56585603
|
0.262
|
NM_001193623
|
LOC147646
|
hypothetical protein LOC147646
|
|
chr16_+_29597965
|
0.261
|
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr3_+_9933763
|
0.261
|
NM_032732
NM_153460
NM_153461
|
IL17RC
|
interleukin 17 receptor C
|
|
chr5_+_75734759
|
0.260
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr12_+_55808696
|
0.259
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr10_+_88506320
|
0.259
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr22_+_38903825
|
0.259
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr10_-_91394881
|
0.259
|
|
PANK1
|
pantothenate kinase 1
|
|
chr6_-_159340737
|
0.258
|
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas)
|
|
chrX_+_48529905
|
0.258
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr2_-_220144511
|
0.258
|
NM_001173408
NM_001173431
NM_015311
|
OBSL1
|
obscurin-like 1
|
|
chr11_-_70641267
|
0.258
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr7_+_128219307
|
0.257
|
NM_022742
|
CCDC136
|
coiled-coil domain containing 136
|
|
chr19_+_50100843
|
0.257
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr12_+_106236281
|
0.256
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr9_-_79835970
|
0.256
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr4_+_144477432
|
0.255
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr5_+_137829077
|
0.255
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr1_+_90060131
|
0.255
|
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr2_-_36384
|
0.254
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr16_-_49742008
|
0.254
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr14_-_37133927
|
0.254
|
|
FOXA1
|
forkhead box A1
|
|
chr11_+_123600553
|
0.253
|
NM_001007249
|
OR8G2
|
olfactory receptor, family 8, subfamily G, member 2
|
|
chr2_-_38683119
|
0.252
|
|
HNRPLL
|
heterogeneous nuclear ribonucleoprotein L-like
|
|
chr1_-_151855413
|
0.251
|
NM_020672
|
S100A14
|
S100 calcium binding protein A14
|
|
chr10_+_17726129
|
0.250
|
NM_003473
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1
|
|
chr17_-_35017691
|
0.250
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr6_-_159340784
|
0.249
|
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas)
|
|
chr1_-_150070849
|
0.248
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr3_+_125786195
|
0.247
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr11_-_32413486
|
0.247
|
|
WT1
|
Wilms tumor 1
|
|
chr14_-_23686269
|
0.246
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr16_+_28785087
|
0.246
|
|
SH2B1
|
SH2B adaptor protein 1
|
|
chr10_-_21854616
|
0.245
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr17_-_39556483
|
0.245
|
|
HDAC5
|
histone deacetylase 5
|
|
chr1_+_3679168
|
0.245
|
NM_001163724
|
LOC388588
|
hypothetical protein LOC388588
|
|
chr17_+_44429748
|
0.244
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr13_+_92677012
|
0.244
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr1_+_57093030
|
0.243
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr12_+_55808514
|
0.243
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr16_+_29597829
|
0.240
|
NM_014298
|
QPRT
|
quinolinate phosphoribosyltransferase
|
|
chr17_-_35017650
|
0.238
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr3_-_159306532
|
0.237
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr12_+_54700955
|
0.237
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr12_+_120726317
|
0.237
|
|
SETD1B
|
SET domain containing 1B
|
|
chr11_+_45900731
|
0.236
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr7_+_137795719
|
0.235
|
|
TRIM24
|
tripartite motif containing 24
|
|
chr20_-_22512893
|
0.234
|
|
FOXA2
|
forkhead box A2
|
|
chr6_+_150112549
|
0.231
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr5_-_134397470
|
0.231
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr17_-_75385371
|
0.231
|
NM_020649
|
CBX8
|
chromobox homolog 8
|
|
chr17_-_62671738
|
0.231
|
|
HELZ
|
helicase with zinc finger
|
|
chr16_+_15644624
|
0.230
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr7_-_100647585
|
0.230
|
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr1_-_166172901
|
0.229
|
NM_001143674
|
BRP44
|
brain protein 44
|
|
chr1_-_143706300
|
0.229
|
NM_001002812
NM_014644
NM_001002810
NM_001198834
NM_001195261
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr13_-_49597776
|
0.229
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr18_+_3439972
|
0.228
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_227544836
|
0.227
|
|
C1orf96
|
chromosome 1 open reading frame 96
|
|
chr16_+_28741908
|
0.226
|
NM_007245
NM_145714
NM_148414
NM_148415
NM_148416
|
ATXN2L
|
ataxin 2-like
|
|
chr14_-_60260211
|
0.225
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr20_-_33335919
|
0.225
|
NM_181468
|
EIF6
|
eukaryotic translation initiation factor 6
|
|
chr1_-_227545310
|
0.223
|
NM_145257
|
C1orf96
|
chromosome 1 open reading frame 96
|
|
chr16_+_29582069
|
0.223
|
NM_003123
|
SPN
|
sialophorin
|
|
chr12_+_48303607
|
0.221
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr19_-_10540328
|
0.221
|
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4)
|
|
chr14_-_20636337
|
0.220
|
|
ZNF219
|
zinc finger protein 219
|
|
chr11_+_46679140
|
0.219
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chr1_+_201862548
|
0.218
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr11_-_27450808
|
0.218
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr6_+_41863386
|
0.218
|
|
TOMM6
|
translocase of outer mitochondrial membrane 6 homolog (yeast)
|
|
chr18_+_30812205
|
0.217
|
NM_001143827
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr6_-_70563489
|
0.217
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr16_+_30667120
|
0.217
|
NM_000294
NM_001172432
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis)
|