|
chr13_+_112999590
|
3.013
|
|
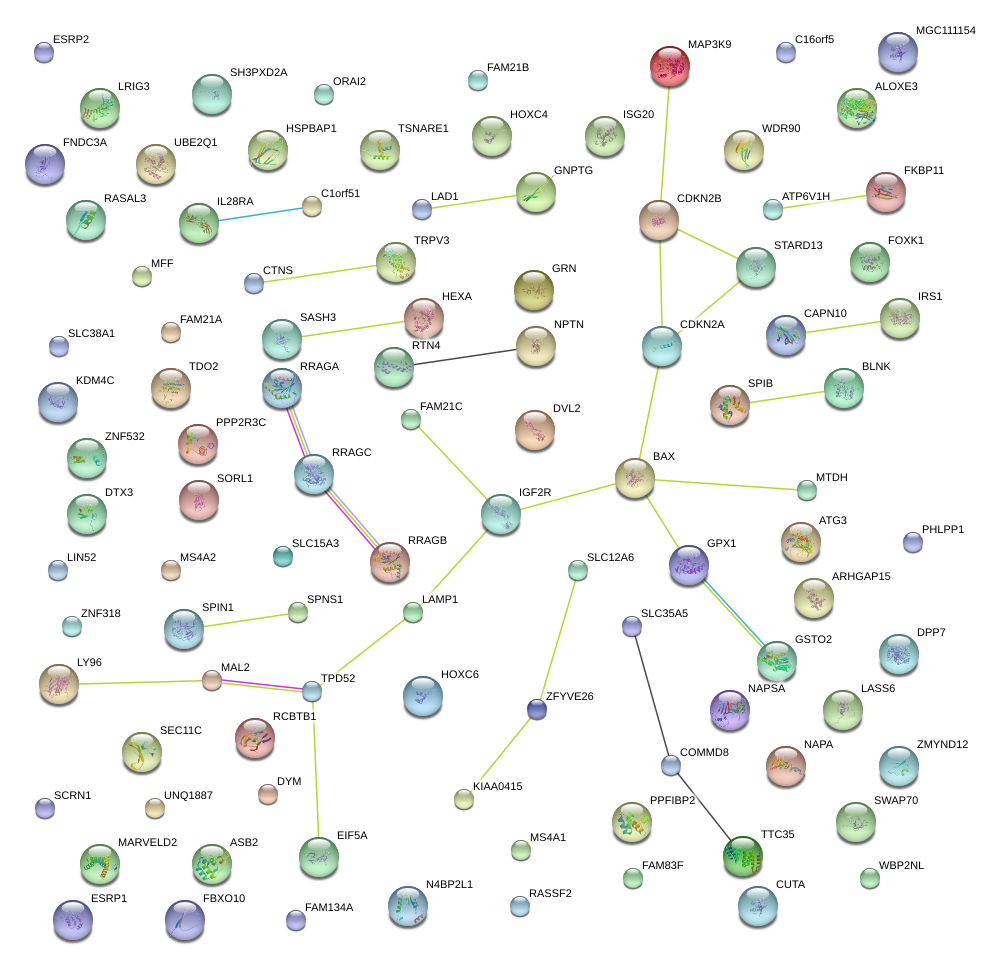
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_112999538
|
2.963
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr13_+_112999462
|
2.596
|
NM_005561
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr19_-_52710129
|
2.094
|
NM_003827
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr12_-_47604930
|
2.090
|
NM_001143781
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr13_+_48582389
|
1.990
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr17_-_7078446
|
1.931
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_-_52709975
|
1.890
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr14_-_67353046
|
1.860
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr19_-_52710089
|
1.782
|
|
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha
|
|
chr14_-_93512818
|
1.760
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr17_-_7078304
|
1.725
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr17_-_7078399
|
1.702
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr15_-_71712711
|
1.702
|
NM_001161363
NM_001161364
NM_012428
NM_017455
|
NPTN
|
neuroplastin
|
|
chr18_+_54958095
|
1.634
|
NM_033280
|
SEC11C
|
SEC11 homolog C (S. cerevisiae)
|
|
chr13_-_31900218
|
1.608
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr19_+_54149963
|
1.587
|
|
BAX
|
BCL2-associated X protein
|
|
chr8_-_54918389
|
1.586
|
NM_015941
NM_213619
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr8_-_54918325
|
1.579
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr10_-_98021144
|
1.576
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chrX_+_55760774
|
1.573
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr19_+_54149941
|
1.571
|
|
BAX
|
BCL2-associated X protein
|
|
chr8_-_54918369
|
1.569
|
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr15_-_71712605
|
1.546
|
|
NPTN
|
neuroplastin
|
|
chr16_+_28893525
|
1.537
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr17_-_3407780
|
1.508
|
|
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3
|
|
chr15_-_70455355
|
1.504
|
NM_000520
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr6_+_160310118
|
1.497
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr8_-_54918099
|
1.455
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr19_+_54149843
|
1.439
|
NM_004324
NM_138761
NM_138763
NM_138764
|
BAX
|
BCL2-associated X protein
|
|
chr13_-_49057707
|
1.400
|
NM_018191
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr13_-_49057629
|
1.399
|
|
RCBTB1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr12_+_52733927
|
1.368
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chrX_-_114703273
|
1.357
|
|
|
|
|
chr17_+_75851984
|
1.353
|
|
|
|
|
chr2_+_143603439
|
1.351
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr8_+_120289735
|
1.292
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr15_-_70455153
|
1.274
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr11_+_120828058
|
1.272
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_+_148521852
|
1.269
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr1_-_199635057
|
1.236
|
|
LAD1
|
ladinin 1
|
|
chr9_+_6747919
|
1.219
|
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr11_+_7491789
|
1.210
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr19_-_15436333
|
1.206
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr11_+_120828275
|
1.205
|
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr2_+_241174803
|
1.204
|
NM_023083
NM_023085
|
CAPN10
|
calpain 10
|
|
chr16_-_66827449
|
1.200
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr11_+_7491576
|
1.176
|
NM_003621
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr17_+_39778016
|
1.164
|
NM_002087
|
GRN
|
granulin
|
|
chr10_-_105427898
|
1.143
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr7_+_4781784
|
1.141
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr14_+_73621408
|
1.139
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr22_+_40724674
|
1.129
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr7_-_29995809
|
1.121
|
NM_001145515
NM_014766
NM_001145513
|
SCRN1
|
secernin 1
|
|
chr16_+_639278
|
1.100
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr1_-_152797760
|
1.090
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr2_+_219751207
|
1.088
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr1_-_42694263
|
1.077
|
|
ZMYND12
|
zinc finger, MYND-type containing 12
|
|
chr1_-_152797707
|
1.075
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr8_+_98725604
|
1.069
|
|
MTDH
|
metadherin
|
|
chr11_+_9642226
|
1.067
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr17_+_7152417
|
1.066
|
NM_001143762
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr17_+_39778153
|
1.060
|
|
GRN
|
granulin
|
|
chr15_-_32398220
|
1.054
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr6_-_33493640
|
1.053
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr2_+_169021001
|
1.052
|
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr12_-_119826073
|
1.051
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr2_+_219751192
|
1.045
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr11_+_9642200
|
1.044
|
NM_015055
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chrX_+_128741578
|
1.043
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr14_-_70345485
|
1.039
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr8_+_109525050
|
1.037
|
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr17_+_3486753
|
1.030
|
|
CTNS
|
cystinosis, nephropathic
|
|
chr2_+_219751230
|
1.028
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr11_+_9642182
|
1.028
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr8_-_81246215
|
1.026
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr15_+_86983038
|
1.026
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr12_-_44949046
|
1.016
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr17_+_39778191
|
1.011
|
|
GRN
|
granulin
|
|
chr6_-_43445103
|
1.010
|
NM_014345
|
ZNF318
|
zinc finger protein 318
|
|
chr1_-_24386324
|
1.008
|
NM_170743
NM_173064
NM_173065
|
IL28RA
|
interleukin 28 receptor, alpha (interferon, lambda receptor)
|
|
chr1_-_152797767
|
1.002
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr8_+_75066117
|
0.999
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr8_+_98725505
|
0.995
|
NM_178812
|
MTDH
|
metadherin
|
|
chr19_+_55614006
|
0.992
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr3_-_113763449
|
0.991
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr2_+_169021039
|
0.987
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr17_+_39778180
|
0.981
|
|
GRN
|
granulin
|
|
chr3_-_49370755
|
0.976
|
NM_000581
NM_201397
|
GPX1
|
glutathione peroxidase 1
|
|
chr8_+_109525010
|
0.975
|
NM_014673
|
TTC35
|
tetratricopeptide repeat domain 35
|
|
chr20_-_4743768
|
0.967
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chrX_+_128741562
|
0.966
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr1_-_152797820
|
0.960
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr3_-_49370682
|
0.954
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr18_+_54682261
|
0.953
|
|
ZNF532
|
zinc finger protein 532
|
|
chr2_+_227898163
|
0.951
|
|
MFF
|
mitochondrial fission factor
|
|
chr8_-_143482433
|
0.950
|
NM_145003
|
TSNARE1
|
t-SNARE domain containing 1
|
|
chr14_-_93512647
|
0.949
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr3_-_113763454
|
0.944
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr13_-_32757835
|
0.943
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_60475558
|
0.943
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr2_-_55090759
|
0.942
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr3_-_123995216
|
0.942
|
NM_024610
|
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1
|
|
chr7_+_101860958
|
0.934
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr1_-_197173159
|
0.932
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr10_+_106018915
|
0.928
|
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr5_+_68746694
|
0.925
|
NM_001038603
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr1_-_39097904
|
0.925
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr9_-_139128777
|
0.924
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr10_+_51497716
|
0.924
|
|
FAM21A
|
family with sequence similarity 21, member A
|
|
chr9_-_21984329
|
0.913
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr2_-_227372574
|
0.912
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_21984379
|
0.908
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr6_-_33493792
|
0.906
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr3_+_113763561
|
0.902
|
NM_017945
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr4_-_47160476
|
0.901
|
|
COMMD8
|
COMM domain containing 8
|
|
chr11_+_9642229
|
0.900
|
|
SWAP70
|
SWAP switching B-cell complex 70kDa subunit
|
|
chr7_+_101861007
|
0.894
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_-_41018291
|
0.891
|
|
|
|
|
chr12_+_56284867
|
0.889
|
NM_178502
|
DTX3
|
deltex homolog 3 (Drosophila)
|
|
chr11_+_59979857
|
0.885
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr12_-_44948976
|
0.885
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr3_-_113763425
|
0.878
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr9_-_21984414
|
0.878
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr4_-_47160403
|
0.877
|
NM_017845
|
COMMD8
|
COMM domain containing 8
|
|
chr2_+_219751145
|
0.873
|
NM_024293
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr22_+_38720881
|
0.873
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr12_-_57600564
|
0.846
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr12_-_57600336
|
0.832
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr16_-_4528380
|
0.831
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr9_-_37566248
|
0.828
|
NM_012166
|
FBXO10
|
F-box protein 10
|
|
chr8_+_95722448
|
0.824
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr17_-_7962531
|
0.816
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr16_+_1341937
|
0.814
|
|
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit
|
|
chr14_-_34661156
|
0.813
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr18_-_45241014
|
0.811
|
NM_017653
|
DYM
|
dymeclin
|
|
chr18_+_58535145
|
0.809
|
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr14_-_64040072
|
0.809
|
NM_006977
|
ZBTB25
|
zinc finger and BTB domain containing 25
|
|
chr7_+_17304707
|
0.809
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr17_+_1574782
|
0.808
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr12_+_102982365
|
0.806
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr4_+_75393053
|
0.804
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr1_+_152459918
|
0.802
|
NM_014847
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr3_-_113763217
|
0.796
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr3_-_126414252
|
0.781
|
NM_024628
|
SLC12A8
|
solute carrier family 12 (potassium/chloride transporters), member 8
|
|
chr14_+_20580224
|
0.775
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr10_-_105442868
|
0.762
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr2_-_219750975
|
0.757
|
|
C2orf24
|
chromosome 2 open reading frame 24
|
|
chr6_+_138230045
|
0.755
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_-_166359016
|
0.750
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr7_+_4781803
|
0.749
|
|
KIAA0415
|
KIAA0415
|
|
chr14_-_91642703
|
0.749
|
NM_001127696
NM_001127697
NM_001164774
NM_001164776
NM_001164777
NM_001164778
NM_001164779
NM_001164780
NM_001164781
NM_001164782
NM_004993
NM_030660
|
ATXN3
|
ataxin 3
|
|
chr14_+_44622916
|
0.747
|
NM_017922
|
PRPF39
|
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae)
|
|
chr16_+_80370426
|
0.746
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr1_-_115060904
|
0.742
|
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog
|
|
chr12_+_52618815
|
0.741
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr15_-_70455794
|
0.740
|
|
HEXA
|
hexosaminidase A (alpha polypeptide)
|
|
chr19_+_50196526
|
0.734
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr5_+_122138677
|
0.734
|
|
SNX2
|
sorting nexin 2
|
|
chr14_-_34661190
|
0.727
|
NM_017917
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr3_-_113763049
|
0.721
|
NM_022488
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr8_+_98725670
|
0.719
|
|
MTDH
|
metadherin
|
|
chr1_+_111544469
|
0.718
|
|
|
|
|
chr12_+_14409875
|
0.714
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr12_-_51912101
|
0.711
|
NM_000966
|
RARG
|
retinoic acid receptor, gamma
|
|
chr22_+_23153529
|
0.704
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr11_+_125586828
|
0.704
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr14_+_23700261
|
0.699
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr3_+_44878380
|
0.699
|
NM_144638
|
TMEM42
|
transmembrane protein 42
|
|
chr14_-_91642641
|
0.699
|
|
ATXN3
ATXN8
|
ataxin 3
ataxin 8
|
|
chr16_-_15644465
|
0.696
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr16_-_66072477
|
0.695
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr19_+_1018293
|
0.695
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr17_+_6856521
|
0.694
|
NM_001004333
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chr3_-_11736946
|
0.694
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr20_+_43028590
|
0.693
|
|
STK4
|
serine/threonine kinase 4
|
|
chr22_-_36907638
|
0.692
|
NM_001004426
NM_003560
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr6_+_139391511
|
0.690
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr3_+_133619193
|
0.687
|
NM_015268
|
DNAJC13
|
DnaJ (Hsp40) homolog, subfamily C, member 13
|
|
chr17_+_6856676
|
0.684
|
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chr20_+_43028533
|
0.677
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr19_+_1018536
|
0.676
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr9_+_6747636
|
0.674
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr3_+_114733907
|
0.671
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr9_+_114953048
|
0.670
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr14_+_44501083
|
0.669
|
NM_015091
|
FAM179B
|
family with sequence similarity 179, member B
|
|
chr12_+_52713098
|
0.666
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr5_+_122138636
|
0.662
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr3_+_113763601
|
0.660
|
|
SLC35A5
|
solute carrier family 35, member A5
|
|
chr3_+_87359088
|
0.659
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr12_+_56271259
|
0.656
|
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma
|
|
chr17_+_6856697
|
0.655
|
|
RNASEK-C17ORF49
|
RNASEK-C17orf49 read-through transcript
|
|
chr17_+_1574583
|
0.653
|
NM_001163811
|
WDR81
|
WD repeat domain 81
|
|
chr9_-_133574928
|
0.650
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr5_+_122138661
|
0.647
|
|
SNX2
|
sorting nexin 2
|
|
chr7_-_27149750
|
0.646
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr1_+_149437602
|
0.642
|
NM_001135636
NM_001135637
NM_001135638
NM_003557
|
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha
|
|
chr1_+_42694751
|
0.634
|
NM_001077447
NM_024664
|
PPCS
|
phosphopantothenoylcysteine synthetase
|
|
chr16_+_68353662
|
0.632
|
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr11_-_85457469
|
0.630
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|