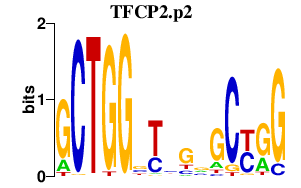
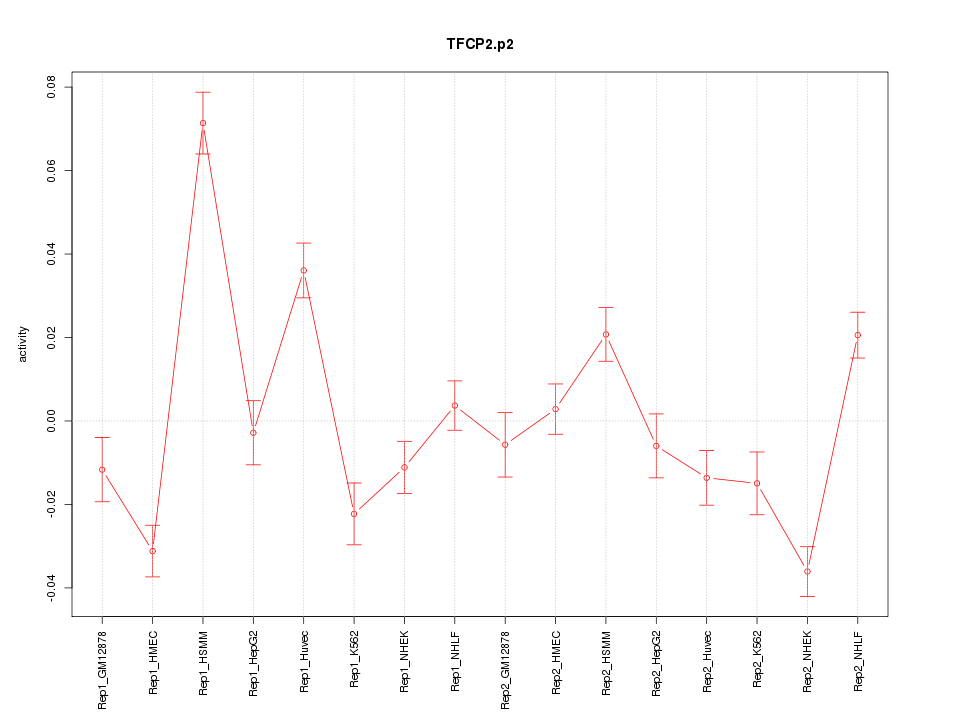
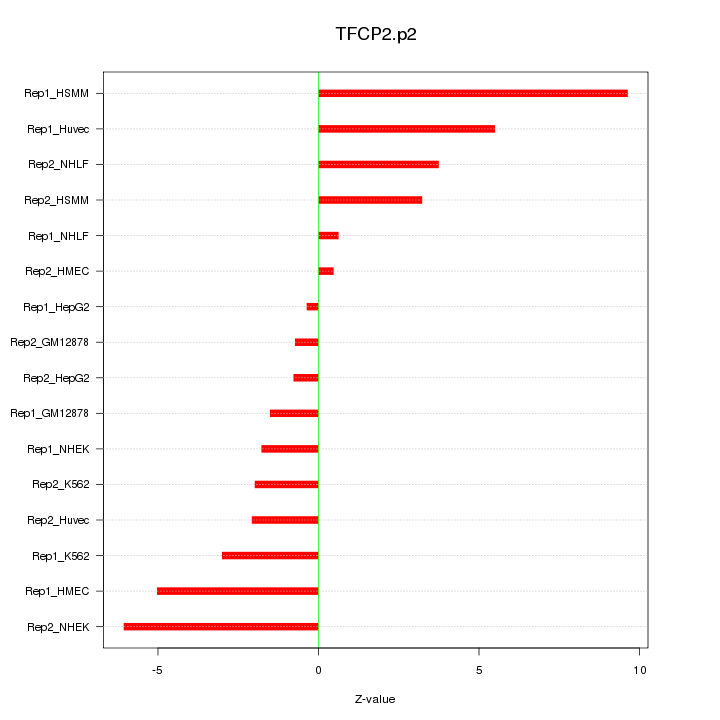
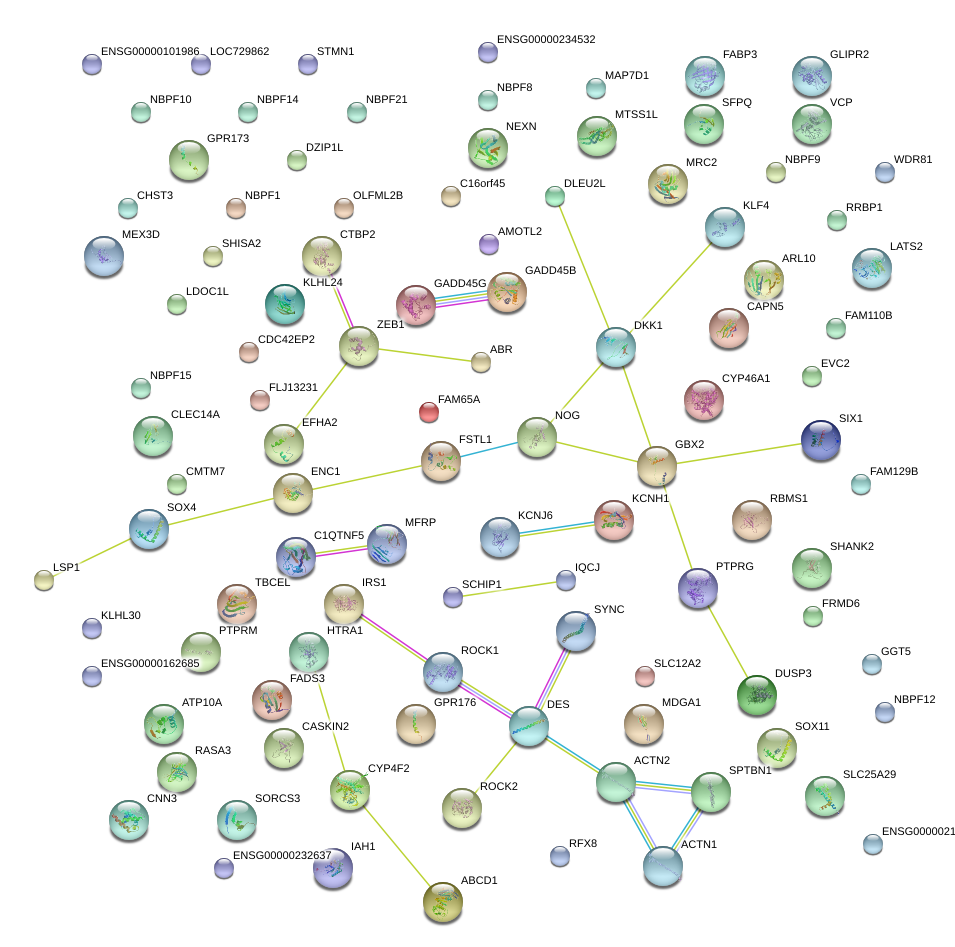
|
chr19_-_1518875
|
4.425
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr3_+_160964574
|
4.050
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_+_238712101
|
3.973
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr1_+_78126787
|
3.667
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr11_+_64838882
|
3.219
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr13_-_20533629
|
3.069
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr19_-_15869870
|
2.935
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr14_-_99842519
|
2.934
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr3_-_121652183
|
2.791
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr18_+_7557313
|
2.752
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr10_+_124210980
|
2.738
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr10_+_31648103
|
2.704
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr9_-_35062643
|
2.689
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr2_-_161058550
|
2.589
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr11_-_61415429
|
2.390
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr10_-_31647925
|
2.388
|
|
LOC220930
|
hypothetical LOC220930
|
|
chr13_-_20533703
|
2.364
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr14_+_51188289
|
2.363
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr11_+_76455574
|
2.336
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr8_+_16929116
|
2.314
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr17_+_52026058
|
2.299
|
NM_005450
|
NOG
|
noggin
|
|
chr2_+_54537842
|
2.282
|
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr11_-_118716736
|
2.226
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr6_-_37773609
|
2.197
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr3_+_184836049
|
2.197
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr2_-_161058120
|
2.180
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr13_-_49597776
|
2.158
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr17_+_58058479
|
2.154
|
NM_006039
|
MRC2
|
mannose receptor, C type 2
|
|
chr11_+_120399966
|
2.152
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr1_-_95165089
|
2.127
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr17_-_71023048
|
2.122
|
|
CASKIN2
|
CASK interacting protein 2
|
|
chr21_-_38210531
|
2.116
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr15_-_37999717
|
2.110
|
|
GPR176
|
G protein-coupled receptor 176
|
|
chr9_+_36126667
|
2.102
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr16_+_66121040
|
2.066
|
NM_001193523
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr14_-_60185659
|
2.061
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr17_-_71023221
|
2.047
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr5_-_37285160
|
1.966
|
NM_023073
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr15_-_23659329
|
1.954
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr10_+_106391032
|
1.926
|
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr17_-_959007
|
1.917
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr19_+_2427122
|
1.916
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr9_-_129371171
|
1.912
|
NM_022833
|
FAM129B
|
family with sequence similarity 129, member B
|
|
chr9_+_36126710
|
1.904
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr15_-_37999519
|
1.875
|
|
|
|
|
chr5_+_175725050
|
1.847
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr14_-_37795001
|
1.832
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr9_-_109291866
|
1.832
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr3_-_135575925
|
1.804
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_-_118716244
|
1.803
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr13_-_113916066
|
1.790
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr11_+_1848674
|
1.783
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr13_-_113916186
|
1.770
|
NM_007368
|
RASA3
|
RAS p21 protein activator 3
|
|
chr5_-_73972055
|
1.770
|
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr17_-_39211706
|
1.762
|
NM_004090
|
DUSP3
|
dual specificity phosphatase 3
|
|
chr1_-_26105477
|
1.762
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chrX_+_53095230
|
1.760
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr13_-_113916159
|
1.740
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr9_+_91409746
|
1.733
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr8_+_59069522
|
1.730
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr3_+_61522922
|
1.727
|
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr13_-_113916115
|
1.723
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr3_+_148593911
|
1.722
|
|
|
|
|
chr9_-_109291575
|
1.713
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr2_+_219991342
|
1.697
|
NM_001927
|
DES
|
desmin
|
|
chr5_+_127447306
|
1.687
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr2_-_11402150
|
1.674
|
NM_004850
|
ROCK2
|
Rho-associated, coiled-coil containing protein kinase 2
|
|
chr1_-_160260210
|
1.668
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr2_+_5750229
|
1.664
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chrX_+_152644385
|
1.661
|
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr13_-_25523164
|
1.654
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr2_-_101457596
|
1.648
|
NM_001145664
|
RFX8
|
regulatory factor X, 8
|
|
chr1_-_32940850
|
1.645
|
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr1_-_31618405
|
1.642
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr22_-_22970850
|
1.630
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr2_+_9532087
|
1.629
|
NM_001039613
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chr2_-_236741352
|
1.625
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr14_+_99220349
|
1.617
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr10_-_126839546
|
1.614
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr16_-_69277355
|
1.612
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr3_+_121296290
|
1.603
|
|
|
|
|
chr11_-_70641267
|
1.594
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_-_227371698
|
1.547
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_35062596
|
1.534
|
|
VCP
|
valosin containing protein
|
|
chr3_+_32408346
|
1.532
|
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr1_+_36394044
|
1.530
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr22_-_43271843
|
1.528
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr1_-_209373937
|
1.526
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr14_-_68515791
|
1.518
|
NM_001102
NM_001130004
NM_001130005
|
ACTN1
|
actinin, alpha 1
|
|
chr20_-_17610828
|
1.514
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr10_+_53744043
|
1.514
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr3_-_139317140
|
1.514
|
NM_173543
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr17_+_1580504
|
1.503
|
|
WDR81
|
WD repeat domain 81
|
|
chr10_+_73393998
|
1.480
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr16_+_15435825
|
1.473
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr12_+_13045702
|
1.470
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr10_+_31647425
|
1.447
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr4_-_5761194
|
1.445
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr1_+_143304983
|
1.443
|
|
NBPF10
NBPF1
|
neuroblastoma breakpoint family, member 10
neuroblastoma breakpoint family, member 1
|
|
chr1_-_35431329
|
1.430
|
NM_005066
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr9_-_35062545
|
1.426
|
|
VCP
|
valosin containing protein
|
|
chr11_-_86061003
|
1.423
|
NM_006680
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr8_+_37672427
|
1.419
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr6_+_108988761
|
1.415
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr17_+_58058774
|
1.411
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr5_-_138758783
|
1.409
|
NM_001161546
|
LOC389333
|
hypothetical protein LOC389333
|
|
chr1_-_33420213
|
1.399
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr4_+_37655385
|
1.398
|
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr16_-_31390926
|
1.389
|
|
|
|
|
chr6_-_134538526
|
1.387
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr7_+_128257698
|
1.385
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr9_-_35062578
|
1.381
|
|
VCP
|
valosin containing protein
|
|
chr14_-_68515731
|
1.377
|
|
ACTN1
|
actinin, alpha 1
|
|
chr9_+_138341752
|
1.374
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr18_-_3001944
|
1.371
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr9_+_96806235
|
1.358
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr9_+_136673534
|
1.358
|
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr1_-_95165037
|
1.351
|
|
CNN3
|
calponin 3, acidic
|
|
chr2_+_85213884
|
1.350
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr17_-_15104768
|
1.347
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr18_-_71049977
|
1.346
|
|
|
|
|
chr16_-_21439195
|
1.345
|
|
SLC7A5P2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 pseudogene 2
|
|
chr1_-_925332
|
1.339
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr1_-_232811846
|
1.339
|
NM_001077397
NM_182972
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr13_-_22847620
|
1.336
|
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chrX_-_106846924
|
1.332
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_-_39871226
|
1.324
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr3_+_129690735
|
1.317
|
|
|
|
|
chr19_-_12266592
|
1.317
|
NM_001164276
NM_016264
|
ZNF44
|
zinc finger protein 44
|
|
chr1_-_35431318
|
1.312
|
|
SFPQ
|
splicing factor proline/glutamine-rich
|
|
chr19_+_4229597
|
1.307
|
NM_020209
|
SHD
|
Src homology 2 domain containing transforming protein D
|
|
chr2_+_71547325
|
1.305
|
NM_001130455
NM_001130982
NM_001130983
NM_001130984
NM_001130985
NM_001130986
NM_001130987
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr14_+_95575319
|
1.302
|
|
C14orf132
|
chromosome 14 open reading frame 132
|
|
chr17_+_58058783
|
1.301
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr2_+_54536780
|
1.301
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr13_-_109236897
|
1.295
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr14_-_68515742
|
1.295
|
|
ACTN1
|
actinin, alpha 1
|
|
chr2_+_200879737
|
1.284
|
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr19_-_9790502
|
1.281
|
|
FBXL12
|
F-box and leucine-rich repeat protein 12
|
|
chr11_+_32993973
|
1.276
|
NM_001077242
|
DEPDC7
|
DEP domain containing 7
|
|
chr11_-_86060812
|
1.275
|
NM_001014811
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr3_+_132583204
|
1.266
|
NM_001171905
NM_001171906
NM_152395
|
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr9_+_81377289
|
1.262
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr13_+_75108450
|
1.261
|
|
LMO7
|
LIM domain 7
|
|
chr5_-_32348826
|
1.248
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr10_+_124211353
|
1.246
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_+_49034050
|
1.245
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr15_-_81667773
|
1.244
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr11_-_2116048
|
1.243
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_+_32408155
|
1.239
|
NM_138410
NM_181472
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr22_+_40800193
|
1.238
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr19_-_12007485
|
1.232
|
|
ZNF433
|
zinc finger protein 433
|
|
chr7_-_525336
|
1.229
|
|
PDGFA
|
platelet-derived growth factor alpha polypeptide
|
|
chr3_-_180651874
|
1.227
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr17_+_1574782
|
1.222
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr19_+_12036513
|
1.221
|
NM_001136501
|
ZNF844
|
zinc finger protein 844
|
|
chr5_-_133589704
|
1.220
|
NM_002715
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr19_-_12007503
|
1.218
|
NM_001080411
|
ZNF433
|
zinc finger protein 433
|
|
chr2_+_200879656
|
1.203
|
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr1_-_32940943
|
1.202
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr12_-_74764677
|
1.201
|
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr16_+_66153810
|
1.194
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr20_-_39379533
|
1.192
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr17_-_73867748
|
1.187
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr17_+_1574583
|
1.187
|
NM_001163811
|
WDR81
|
WD repeat domain 81
|
|
chr16_+_2025922
|
1.186
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr17_-_4216359
|
1.178
|
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast)
|
|
chr2_-_136350404
|
1.175
|
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr14_-_68515761
|
1.167
|
|
ACTN1
|
actinin, alpha 1
|
|
chrX_+_66683075
|
1.166
|
|
AR
|
androgen receptor
|
|
chr7_+_28415495
|
1.166
|
|
LOC401317
|
hypothetical LOC401317
|
|
chr17_+_21128625
|
1.165
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr19_-_11169184
|
1.163
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr19_-_14089362
|
1.163
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr4_+_148872902
|
1.158
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr2_+_71534260
|
1.156
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr2_-_216944911
|
1.156
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr8_-_144969529
|
1.154
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr19_+_40322231
|
1.152
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr10_+_134201262
|
1.151
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr20_+_61622545
|
1.148
|
NM_024299
|
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor homolog (zebrafish)
|
|
chr9_+_89530943
|
1.147
|
|
CTSL1
|
cathepsin L1
|
|
chr8_+_63324053
|
1.145
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr2_-_175578156
|
1.143
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr20_-_17610704
|
1.141
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_-_39124124
|
1.133
|
NM_031899
|
GORASP1
|
golgi reassembly stacking protein 1, 65kDa
|
|
chr19_+_23091616
|
1.128
|
|
ZNF730
|
zinc finger protein 730
|
|
chr16_-_2521359
|
1.125
|
NM_001048212
|
CEMP1
|
cementum protein 1
|
|
chr12_+_104025686
|
1.124
|
|
|
|
|
chr11_+_32994303
|
1.121
|
NM_139160
|
DEPDC7
|
DEP domain containing 7
|
|
chr6_+_147871477
|
1.120
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr20_+_3817593
|
1.118
|
NM_153638
|
PANK2
|
pantothenate kinase 2
|
|
chr7_+_27191534
|
1.117
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr5_-_172594691
|
1.113
|
NM_001166175
NM_001166176
NM_004387
|
NKX2-5
|
NK2 transcription factor related, locus 5 (Drosophila)
|
|
chr8_+_144751216
|
1.105
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr1_+_44185227
|
1.099
|
|
IPO13
|
importin 13
|
|
chr19_+_46417099
|
1.098
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_-_13445942
|
1.094
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr14_-_68515701
|
1.090
|
|
ACTN1
|
actinin, alpha 1
|
|
chr1_+_44185169
|
1.090
|
|
IPO13
|
importin 13
|