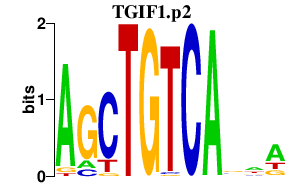
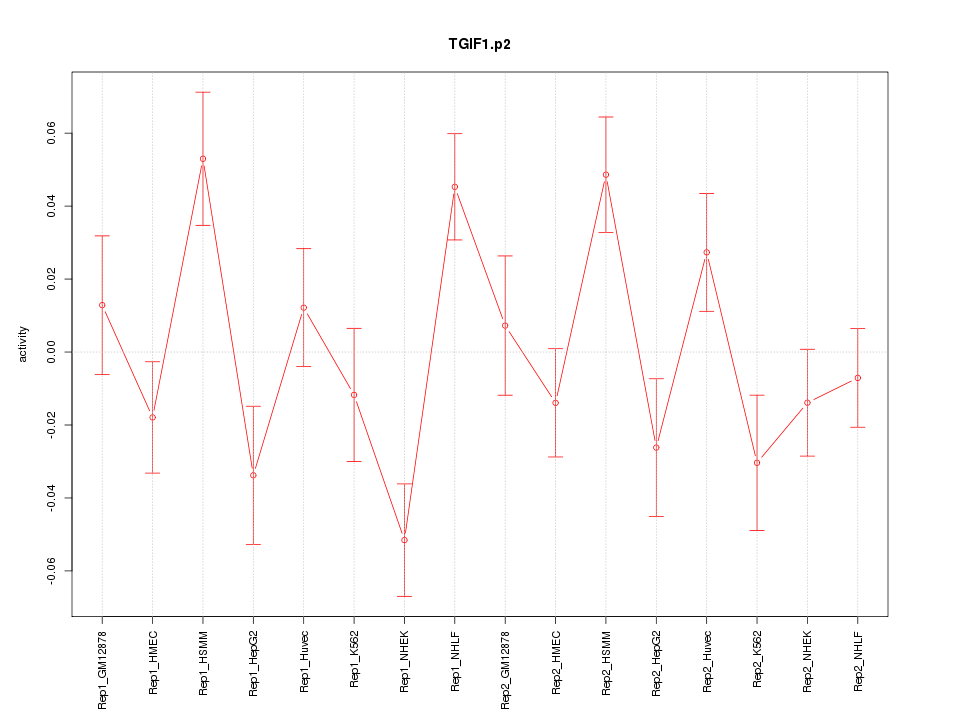
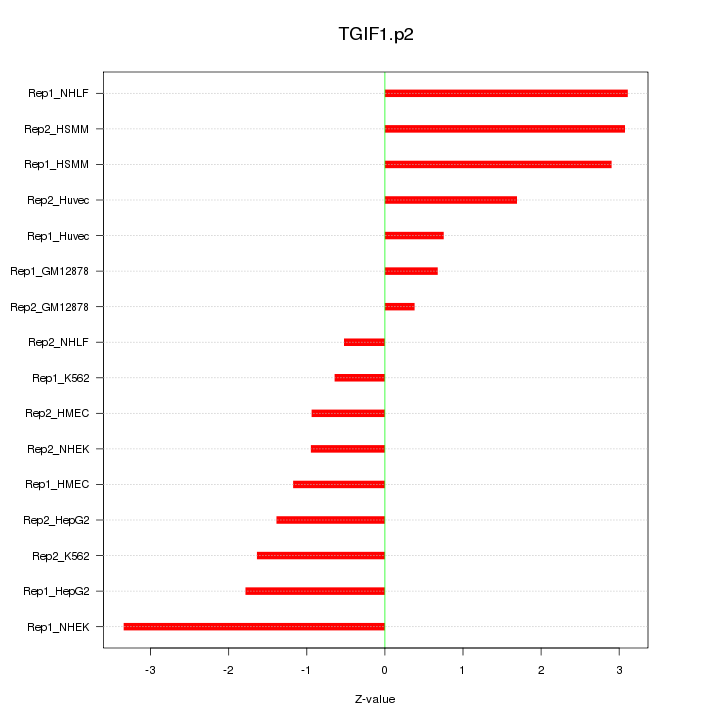
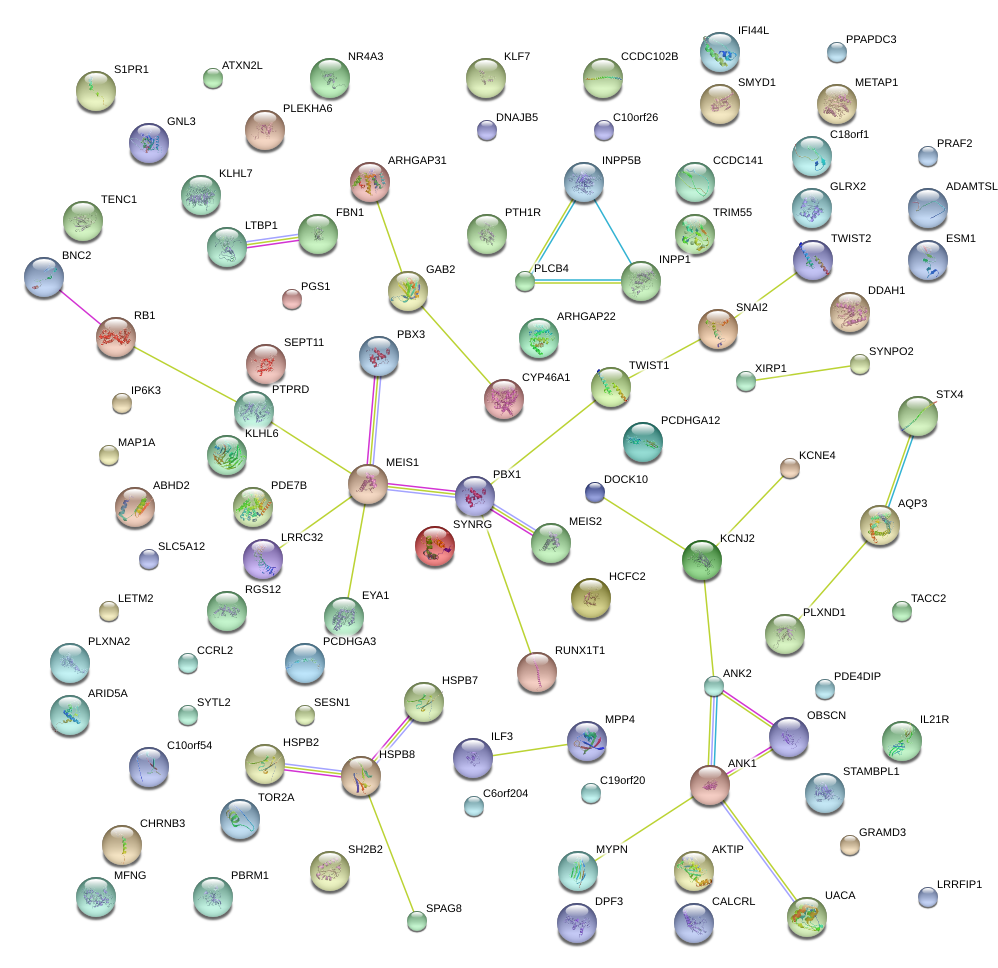
|
chr16_+_27320983
|
3.016
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr15_-_46725087
|
2.599
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr6_-_33822659
|
2.478
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr2_+_33213111
|
2.444
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr15_-_68781662
|
2.393
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr2_+_223625105
|
2.344
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr8_+_67201684
|
2.295
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr2_-_225519974
|
2.227
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr1_-_38185312
|
2.182
|
NM_005540
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr17_+_65677255
|
2.115
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr9_-_16717887
|
2.052
|
|
BNC2
|
basonuclin 2
|
|
chr8_-_41641881
|
2.047
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr1_-_38185307
|
1.966
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr1_-_85816520
|
1.923
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_+_3341521
|
1.878
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr2_+_88148413
|
1.853
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr2_-_202271598
|
1.844
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr2_-_179623030
|
1.841
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr1_-_38185291
|
1.794
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr1_-_38185254
|
1.744
|
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr8_-_49996353
|
1.713
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr2_+_96566190
|
1.647
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr10_+_123862430
|
1.571
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_-_26700121
|
1.568
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr9_+_133154896
|
1.474
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr12_+_51728999
|
1.457
|
NM_015319
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr1_+_101474892
|
1.402
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr14_+_99220349
|
1.393
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr11_-_85115105
|
1.376
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr5_+_125786975
|
1.372
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr11_-_77806413
|
1.371
|
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr12_+_51729239
|
1.366
|
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr10_+_69539255
|
1.361
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_101475149
|
1.349
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr8_+_42671718
|
1.342
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr10_+_90629880
|
1.328
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr1_-_143750988
|
1.316
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr20_+_8997791
|
1.293
|
|
PLCB4
|
phospholipase C, beta 4
|
|
chr5_+_125786717
|
1.247
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr10_-_73203202
|
1.244
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr9_+_101623957
|
1.235
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr16_+_30951915
|
1.233
|
|
STX4
|
syntaxin 4
|
|
chr3_+_120495906
|
1.224
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr9_+_34980715
|
1.224
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr12_+_118100977
|
1.222
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr22_-_36212160
|
1.207
|
NM_001166343
NM_002405
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr10_-_49483002
|
1.176
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr11_-_77806500
|
1.157
|
NM_080491
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr1_-_197173159
|
1.128
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr1_-_206484258
|
1.106
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr2_+_190916863
|
1.093
|
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr10_+_104525877
|
1.072
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr7_+_101715137
|
1.069
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr2_-_188021117
|
1.068
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr15_+_41597097
|
1.052
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr1_-_16217025
|
1.047
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr15_+_87540238
|
1.042
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr11_-_76059435
|
1.041
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr9_+_127550298
|
1.035
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr8_-_93099040
|
1.031
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_72430545
|
1.027
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr3_-_39209075
|
1.022
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr2_+_66519949
|
1.020
|
|
MEIS1
|
Meis homeobox 1
|
|
chr5_+_180190585
|
1.011
|
|
LOC729678
|
hypothetical LOC729678
|
|
chr7_+_101715110
|
1.010
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr6_-_119137923
|
1.008
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr7_-_27158726
|
1.005
|
|
|
|
|
chr4_+_78089968
|
0.983
|
|
SEPT11
|
septin 11
|
|
chr3_+_46423682
|
0.981
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr3_-_52694554
|
0.972
|
NM_018313
|
PBRM1
|
polybromo 1
|
|
chr2_+_238200922
|
0.965
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr6_-_109521969
|
0.952
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr8_+_38363150
|
0.949
|
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr3_-_130807865
|
0.940
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr2_-_207738858
|
0.919
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr9_+_18464090
|
0.917
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr5_+_180190542
|
0.909
|
|
|
|
|
chr18_+_64616296
|
0.905
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr9_+_18464138
|
0.898
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr5_+_140848905
|
0.895
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr9_-_35802142
|
0.893
|
NM_001039592
NM_172312
|
SPAG8
|
sperm associated antigen 8
|
|
chr6_+_136214495
|
0.889
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chrX_-_48818585
|
0.887
|
NM_007213
|
PRAF2
|
PRA1 domain family, member 2
|
|
chr3_-_184756101
|
0.886
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr12_+_102982365
|
0.882
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr5_-_54317102
|
0.873
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr1_-_202595666
|
0.873
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr3_+_46894239
|
0.870
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr4_+_120168002
|
0.862
|
|
SYNPO2
|
synaptopodin 2
|
|
chr1_+_78858675
|
0.846
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr16_-_52094613
|
0.832
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr9_-_129537372
|
0.826
|
NM_001085347
NM_001134430
NM_001134431
NM_130459
|
TOR2A
|
torsin family 2, member A
|
|
chr4_+_113958652
|
0.818
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr17_-_33043514
|
0.804
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr3_+_52694975
|
0.802
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr1_-_191341866
|
0.800
|
NM_016066
|
GLRX2
|
glutaredoxin 2
|
|
chr11_+_111288669
|
0.796
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr17_+_73886290
|
0.795
|
NM_024419
|
PGS1
|
phosphatidylglycerophosphate synthase 1
|
|
chr1_+_226462363
|
0.790
|
NM_001098623
NM_052843
|
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr4_+_120167950
|
0.789
|
|
SYNPO2
|
synaptopodin 2
|
|
chr9_-_8723893
|
0.789
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr9_-_33437527
|
0.788
|
NM_004925
|
AQP3
|
aquaporin 3 (Gill blood group)
|
|
chr8_-_72431274
|
0.786
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr11_-_76058542
|
0.778
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr4_+_120167770
|
0.775
|
|
SYNPO2
|
synaptopodin 2
|
|
chr7_-_19123765
|
0.765
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr18_+_13601664
|
0.761
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr13_+_47775951
|
0.759
|
|
RB1
|
retinoblastoma 1
|
|
chr20_+_8997660
|
0.755
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr11_+_111288686
|
0.748
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr9_-_71476917
|
0.746
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr7_+_33911636
|
0.737
|
NM_133468
|
BMPER
|
BMP binding endothelial regulator
|
|
chr10_+_94598204
|
0.736
|
NM_019053
|
EXOC6
|
exocyst complex component 6
|
|
chr1_+_36394338
|
0.736
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr10_+_124210980
|
0.733
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr5_-_36337756
|
0.732
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr12_+_8741746
|
0.731
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr4_+_146622154
|
0.730
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr9_-_133605037
|
0.729
|
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr8_-_6407882
|
0.728
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr13_-_59223074
|
0.727
|
|
|
|
|
chr6_-_109522207
|
0.720
|
|
SESN1
|
sestrin 1
|
|
chr8_-_42184194
|
0.720
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr6_+_32515596
|
0.707
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr7_-_137337354
|
0.706
|
NM_194071
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr14_+_92049396
|
0.702
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr11_+_63730720
|
0.701
|
NM_031471
NM_178443
|
FERMT3
|
fermitin family member 3
|
|
chr5_-_172976212
|
0.700
|
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr12_+_76749199
|
0.697
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr4_+_159909658
|
0.690
|
|
FNIP2
|
folliculin interacting protein 2
|
|
chr19_-_54557450
|
0.686
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr6_-_27948065
|
0.685
|
NM_003533
|
HIST1H3I
|
histone cluster 1, H3i
|
|
chr1_-_153098811
|
0.683
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_-_19123787
|
0.682
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr9_+_34980266
|
0.681
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr20_-_49612574
|
0.680
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr9_+_129900637
|
0.680
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr1_-_143643064
|
0.679
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_-_14089362
|
0.679
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr16_-_65774904
|
0.678
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr9_-_36266930
|
0.677
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr5_+_149867866
|
0.676
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr12_+_63958689
|
0.673
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr9_-_131845077
|
0.670
|
|
FNBP1
|
formin binding protein 1
|
|
chr13_+_47776026
|
0.663
|
|
RB1
|
retinoblastoma 1
|
|
chr4_+_54790020
|
0.662
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr2_-_179380301
|
0.660
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr12_+_108921776
|
0.652
|
|
ANKRD13A
|
ankyrin repeat domain 13A
|
|
chr2_+_33514896
|
0.650
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr15_-_48192804
|
0.642
|
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr10_+_102662675
|
0.640
|
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr10_+_70331100
|
0.640
|
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50
|
|
chr10_+_115300579
|
0.640
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr19_+_53664386
|
0.632
|
|
CYTH2
|
cytohesin 2
|
|
chr5_+_149845503
|
0.631
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr9_+_129900590
|
0.626
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr5_-_159730196
|
0.626
|
NM_031908
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2
|
|
chr9_-_131845277
|
0.626
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr19_-_14089560
|
0.621
|
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr1_-_171442937
|
0.618
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_-_112301123
|
0.607
|
NM_002037
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr9_+_17124974
|
0.606
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr16_-_65775326
|
0.600
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr3_-_129024665
|
0.593
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr4_+_120167721
|
0.591
|
|
SYNPO2
|
synaptopodin 2
|
|
chr13_-_49597776
|
0.583
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr1_+_26220852
|
0.581
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr9_-_35640852
|
0.580
|
NM_014450
|
SIT1
|
signaling threshold regulating transmembrane adaptor 1
|
|
chr19_+_16169664
|
0.578
|
NM_001130524
NM_032493
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr5_-_172976259
|
0.577
|
NM_001159651
NM_138369
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr1_+_149309703
|
0.576
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr9_+_131123115
|
0.574
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr2_+_71059082
|
0.569
|
NM_001115116
NM_024933
|
ANKRD53
|
ankyrin repeat domain 53
|
|
chr10_+_70330961
|
0.569
|
NM_024045
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50
|
|
chr19_-_44518516
|
0.565
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr5_-_172976196
|
0.562
|
|
BOD1
|
biorientation of chromosomes in cell division 1
|
|
chr2_+_170074457
|
0.559
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr15_+_42815851
|
0.557
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr12_-_69289889
|
0.554
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr6_-_56824672
|
0.545
|
|
DST
|
dystonin
|
|
chr15_+_90738108
|
0.545
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr12_+_108921686
|
0.544
|
|
ANKRD13A
|
ankyrin repeat domain 13A
|
|
chr11_-_127897271
|
0.539
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr3_+_114733907
|
0.539
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr12_+_108921616
|
0.535
|
NM_033121
|
ANKRD13A
|
ankyrin repeat domain 13A
|
|
chr3_+_45042754
|
0.535
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr6_+_151229003
|
0.533
|
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr7_+_120378052
|
0.532
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr6_-_46401363
|
0.518
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr13_-_32757835
|
0.518
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr9_-_14769530
|
0.515
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr8_+_94999167
|
0.514
|
NM_001161778
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr4_-_158111916
|
0.512
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr3_+_23219707
|
0.508
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr10_+_102662315
|
0.506
|
NM_001136123
NM_018121
|
FAM178A
|
family with sequence similarity 178, member A
|
|
chr1_+_26220895
|
0.506
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr13_-_22905822
|
0.504
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr19_-_51856129
|
0.503
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr9_-_129580732
|
0.497
|
NM_170600
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr11_+_111353218
|
0.496
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|