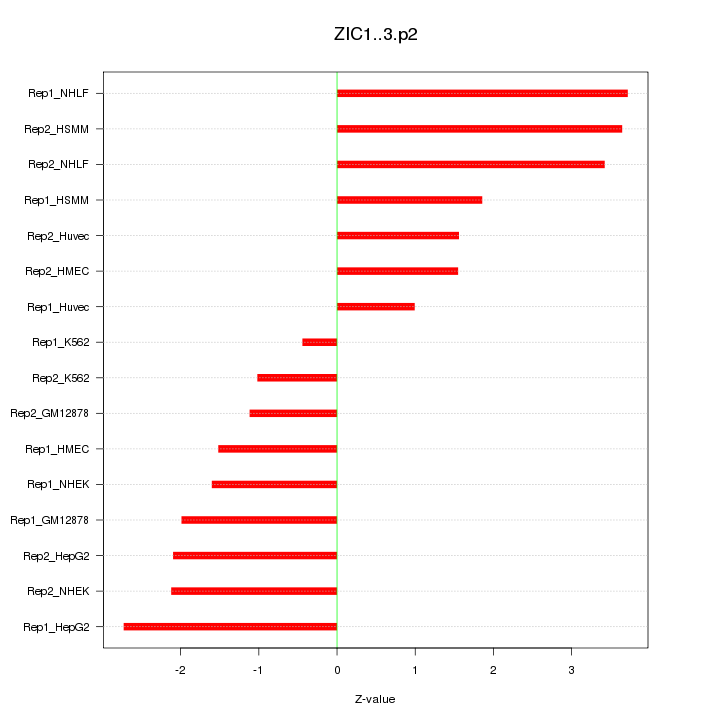
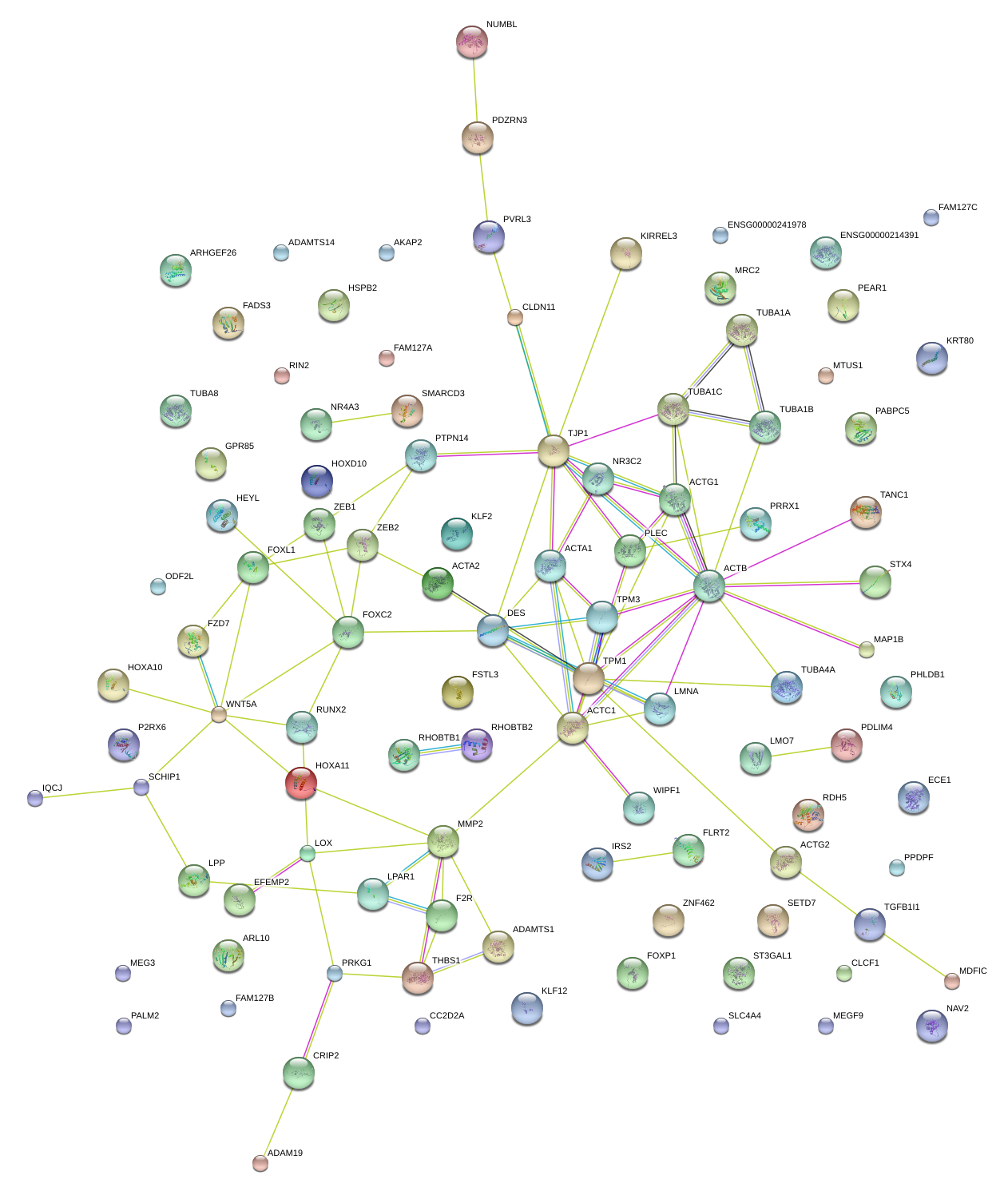
|
chr16_+_85169615
|
6.537
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr3_+_160964574
|
5.312
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr15_+_37660568
|
3.928
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr3_-_73756668
|
3.359
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr11_+_19691397
|
3.258
|
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr4_+_72271218
|
2.941
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr3_+_64645819
|
2.874
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr1_-_21478571
|
2.834
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_-_144991556
|
2.583
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr16_+_85158357
|
2.505
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr10_-_62373828
|
2.503
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr2_-_144991386
|
2.470
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr7_-_27191287
|
2.287
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr9_-_122516399
|
2.228
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr4_-_140696740
|
2.216
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr20_+_19903778
|
2.173
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr9_+_108665168
|
2.075
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr11_+_117983515
|
2.064
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr7_-_112513379
|
2.055
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr21_-_27139143
|
1.998
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr17_+_58058783
|
1.982
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr10_+_52421120
|
1.980
|
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr9_+_111850698
|
1.976
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr21_-_27139563
|
1.947
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr12_-_50871953
|
1.944
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr7_-_106088565
|
1.933
|
NM_175884
|
FLJ36031
|
hypothetical protein FLJ36031
|
|
chr5_-_92982927
|
1.904
|
|
|
|
|
chr11_+_111288669
|
1.893
|
NM_001541
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr9_+_101623957
|
1.890
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr11_+_111288686
|
1.854
|
|
HSPB2-C11ORF52
HSPB2
|
HSPB2-C11orf52 read-through transcript
heat shock 27kDa protein 2
|
|
chr11_-_66898223
|
1.843
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr19_-_45888259
|
1.836
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr16_+_54070302
|
1.798
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_-_39877883
|
1.784
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr9_-_112840064
|
1.780
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr5_+_175725050
|
1.779
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr7_-_150576634
|
1.760
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr15_-_27901828
|
1.752
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr1_-_39877928
|
1.742
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr13_-_73605914
|
1.718
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr8_-_145097031
|
1.716
|
NM_201380
|
PLEC
|
plectin
|
|
chr19_+_16296632
|
1.601
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_-_65396775
|
1.584
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr3_-_71196696
|
1.578
|
|
FOXP1
|
forkhead box P1
|
|
chr2_+_159533329
|
1.557
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr7_-_27180398
|
1.546
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr22_+_19699441
|
1.540
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr4_-_149582948
|
1.537
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr8_-_17599438
|
1.535
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr13_+_75108450
|
1.535
|
|
LMO7
|
LIM domain 7
|
|
chr16_-_31390926
|
1.524
|
|
|
|
|
chr10_+_72102563
|
1.521
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr3_+_171619346
|
1.513
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr7_+_27190688
|
1.507
|
|
|
|
|
chr5_+_71438796
|
1.505
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr6_+_45498354
|
1.500
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr11_-_61415429
|
1.498
|
NM_021727
|
FADS3
|
fatty acid desaturase 3
|
|
chr19_+_16296734
|
1.468
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr7_-_150576660
|
1.463
|
NM_001003801
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr5_-_121440704
|
1.452
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr16_+_31390567
|
1.432
|
NM_001042454
NM_001164719
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr5_-_156935283
|
1.429
|
|
|
|
|
chr2_+_202607554
|
1.422
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr7_+_27191534
|
1.395
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr2_+_219991342
|
1.389
|
NM_001927
|
DES
|
desmin
|
|
chrX_+_133993984
|
1.388
|
NM_001078171
|
FAM127A
|
family with sequence similarity 127, member A
|
|
chr8_-_134653194
|
1.373
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr3_-_55490401
|
1.353
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_-_27190882
|
1.342
|
|
HOXA11
|
homeobox A11
|
|
chr15_+_61127367
|
1.338
|
NM_001018008
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr3_+_112273352
|
1.325
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr14_+_104218487
|
1.315
|
|
|
|
|
chr6_+_45498262
|
1.301
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr3_+_155321826
|
1.301
|
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr1_+_168899936
|
1.300
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chrX_+_90576252
|
1.291
|
NM_080832
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5
|
|
chr11_-_126375864
|
1.281
|
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr16_+_30951915
|
1.275
|
|
STX4
|
syntaxin 4
|
|
chr4_-_149582754
|
1.273
|
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr3_+_112273464
|
1.265
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr16_+_30952382
|
1.250
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr4_+_15089931
|
1.243
|
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr1_-_86634511
|
1.238
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chr12_+_54400417
|
1.221
|
NM_002905
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis)
|
|
chr5_-_156935317
|
1.196
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr10_-_90702490
|
1.195
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr16_+_30952350
|
1.194
|
|
STX4
|
syntaxin 4
|
|
chr20_+_61622985
|
1.181
|
|
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor homolog (zebrafish)
|
|
chr3_+_189353502
|
1.179
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr5_+_131621276
|
1.171
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr13_-_109236897
|
1.166
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr7_+_114350144
|
1.163
|
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr1_+_154351082
|
1.148
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr11_-_110917632
|
1.145
|
|
|
|
|
chr7_-_150576957
|
1.143
|
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr1_-_212791188
|
1.134
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr5_+_131621287
|
1.131
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr12_-_47869134
|
1.129
|
|
TUBA1A
|
tubulin, alpha 1a
|
|
chrX_-_134013644
|
1.123
|
NM_001078172
NM_001134321
|
FAM127B
|
family with sequence similarity 127, member B
|
|
chr9_+_111850770
|
1.121
|
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr14_+_100362236
|
1.116
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr1_+_155130146
|
1.115
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr5_+_76047536
|
1.108
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr14_+_85066265
|
1.098
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr15_-_27901628
|
1.098
|
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr19_+_627346
|
1.092
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr14_+_105012106
|
1.092
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr11_-_126375895
|
1.088
|
NM_001161707
NM_032531
|
KIRREL3
|
kin of IRRE like 3 (Drosophila)
|
|
chr2_-_175255717
|
1.087
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_-_85529884
|
1.087
|
|
TBX18
|
T-box 18
|
|
chr12_-_47869067
|
1.086
|
NM_006009
|
TUBA1A
|
tubulin, alpha 1a
|
|
chr12_+_8741746
|
1.081
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr7_+_44110527
|
1.080
|
|
AEBP1
|
AE binding protein 1
|
|
chr13_+_30378311
|
1.075
|
NM_032849
|
C13orf33
|
chromosome 13 open reading frame 33
|
|
chr22_-_18841785
|
1.071
|
NM_015672
|
RIMBP3
|
RIMS binding protein 3
|
|
chr14_+_100362204
|
1.067
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr4_+_123967389
|
1.054
|
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr12_-_3732614
|
1.052
|
NM_001144958
NM_001144959
NM_032680
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr19_-_54557450
|
1.049
|
NM_003598
|
TEAD2
|
TEA domain family member 2
|
|
chr5_+_131621230
|
1.045
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr12_+_95112385
|
1.043
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr12_+_52653176
|
1.043
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr11_-_3035172
|
1.038
|
NM_001014437
NM_001194997
NM_001751
NM_139273
|
CARS
|
cysteinyl-tRNA synthetase
|
|
chr3_-_180652024
|
1.036
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr19_-_51856129
|
1.035
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr12_-_107745455
|
1.033
|
NM_001161331
|
SSH1
|
slingshot homolog 1 (Drosophila)
|
|
chr2_+_200031333
|
1.032
|
|
|
|
|
chr9_+_34980266
|
1.027
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr2_+_130846263
|
1.018
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chrX_+_149282097
|
1.016
|
NM_001177465
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr1_-_151804929
|
1.016
|
NM_005978
|
S100A2
|
S100 calcium binding protein A2
|
|
chr2_-_175578156
|
1.014
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_+_123967265
|
1.011
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr19_-_60573552
|
1.007
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr3_-_107070400
|
0.999
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr11_-_111287657
|
0.997
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr15_+_41597097
|
0.989
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr10_-_105202012
|
0.987
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr2_+_235525355
|
0.986
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr3_-_180651874
|
0.986
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chrX_-_133984157
|
0.982
|
NM_001078173
|
FAM127C
|
family with sequence similarity 127, member C
|
|
chr11_-_122571046
|
0.974
|
|
CLMP
|
CXADR-like membrane protein
|
|
chr17_+_52026058
|
0.970
|
NM_005450
|
NOG
|
noggin
|
|
chr4_+_159311353
|
0.970
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr19_-_13808101
|
0.968
|
|
LOC284454
|
hypothetical LOC284454
|
|
chr12_-_3732478
|
0.966
|
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr12_+_6179736
|
0.964
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr17_+_35751796
|
0.952
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr11_-_66897584
|
0.950
|
NM_013246
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr11_+_122806196
|
0.947
|
|
LOC100128242
|
hypothetical LOC100128242
|
|
chr12_+_2870255
|
0.942
|
NM_001160408
NM_003324
|
TULP3
|
tubby like protein 3
|
|
chr6_+_21701942
|
0.940
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr1_+_17817397
|
0.939
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr17_-_44010543
|
0.936
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr5_-_141684652
|
0.910
|
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr15_-_58477407
|
0.908
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr1_-_204048758
|
0.902
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr17_+_56884560
|
0.900
|
|
TBX4
|
T-box 4
|
|
chr12_+_6517600
|
0.897
|
|
|
|
|
chrX_-_114374860
|
0.892
|
NM_020871
|
LRCH2
|
leucine-rich repeats and calponin homology (CH) domain containing 2
|
|
chr7_-_27120015
|
0.888
|
|
HOXA3
|
homeobox A3
|
|
chr22_+_35777561
|
0.886
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr12_+_120635021
|
0.873
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr1_-_119991912
|
0.873
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr12_-_94466701
|
0.870
|
NM_001042403
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr17_+_45703765
|
0.867
|
NM_001168215
|
TMEM92
|
transmembrane protein 92
|
|
chr1_+_9275518
|
0.866
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr1_-_93919789
|
0.865
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr5_+_140835788
|
0.863
|
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr5_-_83715947
|
0.862
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr11_-_85053709
|
0.861
|
|
CREBZF
|
CREB/ATF bZIP transcription factor
|
|
chr4_+_81406765
|
0.860
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr5_-_141684759
|
0.860
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr19_+_4861339
|
0.859
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr3_+_185536376
|
0.857
|
NM_001171093
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr1_+_9275542
|
0.851
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr13_+_41520888
|
0.850
|
NM_152910
NM_178009
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr6_+_17389752
|
0.847
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr12_+_12935222
|
0.846
|
NM_003979
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr3_-_130807865
|
0.846
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr8_+_26296439
|
0.844
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr21_+_41461962
|
0.842
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr20_-_3713509
|
0.840
|
|
CENPB
|
centromere protein B, 80kDa
|
|
chr16_-_73842884
|
0.839
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr17_+_17816851
|
0.837
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr11_+_45825244
|
0.836
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr11_+_45825532
|
0.835
|
NM_021117
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr21_-_35181094
|
0.834
|
|
|
|
|
chr4_-_54625463
|
0.831
|
|
CHIC2
|
cysteine-rich hydrophobic domain 2
|
|
chr21_-_35181308
|
0.824
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_148788410
|
0.815
|
NM_019032
NM_025008
|
ADAMTSL4
|
ADAMTS-like 4
|
|
chr1_+_181259317
|
0.814
|
|
|
|
|
chr9_+_34979674
|
0.810
|
NM_001135004
NM_012266
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr1_+_31658549
|
0.809
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr17_-_45629401
|
0.807
|
|
|
|
|
chr12_+_95112131
|
0.804
|
NM_005230
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr12_-_21985433
|
0.802
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr12_+_12935619
|
0.802
|
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr2_-_9061193
|
0.795
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr7_+_44110470
|
0.794
|
NM_001129
|
AEBP1
|
AE binding protein 1
|