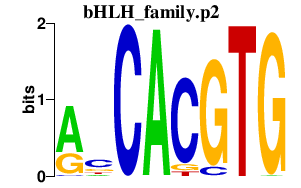
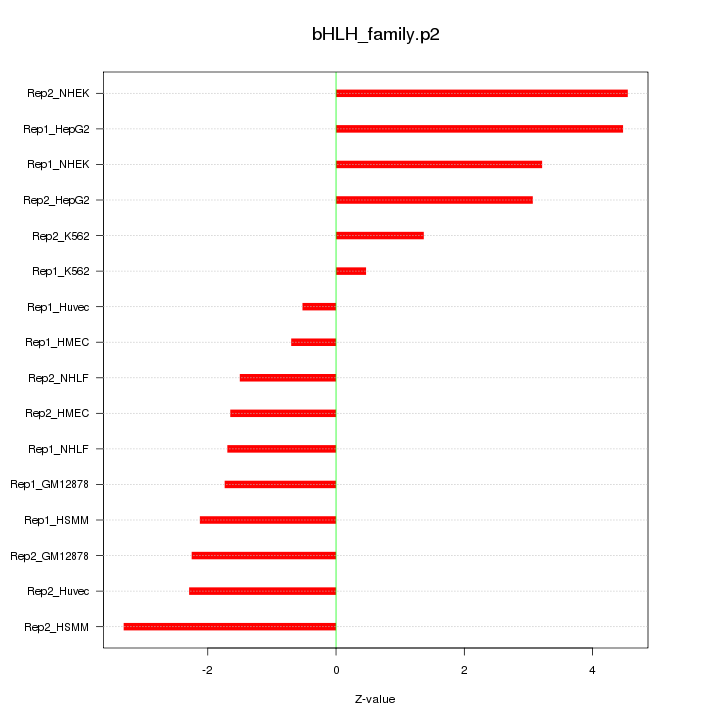
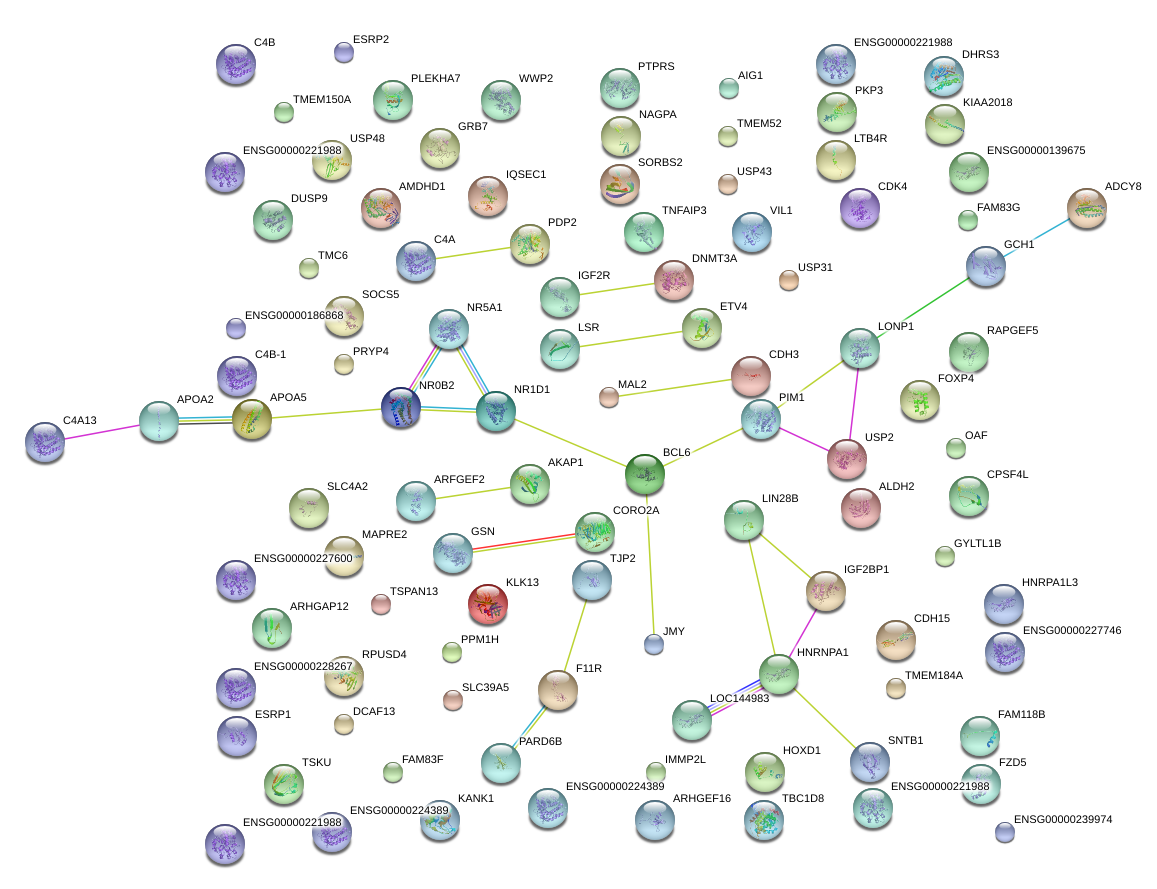
|
chr8_+_120289735
|
4.180
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_-_66827449
|
4.148
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr8_-_17702991
|
3.671
|
|
|
|
|
chr6_+_143423738
|
2.888
|
|
AIG1
|
androgen-induced 1
|
|
chr6_+_143423656
|
2.872
|
NM_016108
|
AIG1
|
androgen-induced 1
|
|
chr16_-_5023934
|
2.659
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr6_+_143423723
|
2.616
|
|
AIG1
|
androgen-induced 1
|
|
chr16_-_23068010
|
2.598
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr16_-_5023914
|
2.572
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr22_+_38720881
|
2.541
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr1_-_12599934
|
2.538
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr12_-_56432292
|
2.516
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr7_-_1562272
|
2.485
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr17_+_9489578
|
2.446
|
NM_153210
|
USP43
|
ubiquitin specific peptidase 43
|
|
chr5_+_78567604
|
2.423
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr12_-_56432375
|
2.363
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432385
|
2.357
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432377
|
2.348
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_73636362
|
2.339
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr2_-_172572993
|
2.283
|
|
|
|
|
chr2_+_172573049
|
2.259
|
NM_199227
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial)
|
|
chr17_-_35510431
|
2.228
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr17_+_35148084
|
2.192
|
NM_001030002
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr9_+_696744
|
2.190
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr17_-_68769613
|
2.171
|
NM_001129885
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like
|
|
chr6_+_138230045
|
2.107
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr16_+_5023672
|
2.084
|
|
LOC100507589
|
hypothetical LOC100507589
|
|
chr20_+_46971618
|
2.076
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr11_-_116168316
|
2.076
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr7_-_22363036
|
2.065
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr11_-_118757477
|
2.057
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_27113018
|
2.057
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr1_-_159459999
|
2.053
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr19_-_5291700
|
2.047
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr16_-_5023562
|
2.011
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr17_+_44429748
|
1.966
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr17_-_18848713
|
1.964
|
NM_001039999
|
FAM83G
|
family with sequence similarity 83, member G
|
|
chr17_-_38979178
|
1.930
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr2_-_208342296
|
1.929
|
NM_003468
|
FZD5
|
frizzled homolog 5 (Drosophila)
|
|
chr7_+_16759845
|
1.923
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr17_-_35510074
|
1.921
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr11_-_125586733
|
1.893
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr12_-_61614930
|
1.873
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr2_+_218992053
|
1.868
|
NM_007127
|
VIL1
|
villin 1
|
|
chr5_+_78568094
|
1.841
|
|
|
|
|
chr17_+_35147738
|
1.836
|
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr17_+_35147697
|
1.812
|
NM_005310
|
GRB7
|
growth factor receptor-bound protein 7
|
|
chr11_+_125586828
|
1.811
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr2_+_46779553
|
1.801
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr12_-_61615124
|
1.780
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr16_+_65471881
|
1.765
|
NM_020786
|
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2
|
|
chr9_-_126309510
|
1.750
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr6_+_41622267
|
1.723
|
|
FOXP4
|
forkhead box P4
|
|
chr7_-_110989582
|
1.690
|
NM_032549
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae)
|
|
chr2_+_176761552
|
1.689
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr14_+_23855291
|
1.669
|
|
LTB4R
|
leukotriene B4 receptor
|
|
chr6_+_160310118
|
1.651
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr7_+_150390566
|
1.648
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr11_+_76171780
|
1.594
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr11_+_384199
|
1.591
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr19_+_40431122
|
1.577
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr6_+_32229771
|
1.569
|
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr6_+_32090516
|
1.559
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr9_-_99994704
|
1.559
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr20_+_48781457
|
1.554
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr2_-_101134146
|
1.537
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr11_+_119586879
|
1.531
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr1_-_1840564
|
1.530
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr6_+_105511615
|
1.521
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr10_-_32257693
|
1.506
|
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr3_-_188945921
|
1.492
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_+_32230172
|
1.487
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr8_+_95722448
|
1.479
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr11_+_384238
|
1.471
|
|
PKP3
|
plakophilin 3
|
|
chr11_-_16992524
|
1.459
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr1_-_159257486
|
1.456
|
|
F11R
|
F11 receptor
|
|
chrX_+_152562013
|
1.450
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr1_-_159257567
|
1.442
|
|
F11R
|
F11 receptor
|
|
chr14_-_54438913
|
1.427
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr17_+_52518098
|
1.426
|
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chr12_+_52960798
|
1.420
|
|
HNRNPA1L2
HNRNPA1
HNRNPA1P10
|
heterogeneous nuclear ribonucleoprotein A1-like 2
heterogeneous nuclear ribonucleoprotein A1
heterogeneous nuclear ribonucleoprotein A1 pseudogene 10
|
|
chr8_-_121893468
|
1.416
|
NM_021021
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr4_-_186970366
|
1.400
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr12_+_94861201
|
1.381
|
NM_152435
|
AMDHD1
|
amidohydrolase domain containing 1
|
|
chr3_-_12983959
|
1.363
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr9_+_70926009
|
1.362
|
NM_001170414
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr1_+_32992131
|
1.348
|
|
|
|
|
chr16_+_68353738
|
1.330
|
NM_007014
NM_199423
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr2_-_25328452
|
1.327
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr18_+_30810889
|
1.320
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr11_+_45900731
|
1.312
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr1_+_3360880
|
1.311
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr12_+_54910767
|
1.309
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr2_-_85683268
|
1.300
|
NM_001031738
|
TMEM150A
|
transmembrane protein 150A
|
|
chr16_+_67236699
|
1.290
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr6_+_37245860
|
1.286
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr2_+_206732859
|
1.273
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr3_-_188946168
|
1.271
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr6_-_53517397
|
1.262
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr11_-_118757574
|
1.257
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr10_-_82039158
|
1.253
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chrX_-_132947290
|
1.251
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr17_-_34084682
|
1.239
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr1_-_21868380
|
1.232
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr19_+_40431359
|
1.231
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr16_+_56616782
|
1.231
|
NM_002428
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr16_+_68353662
|
1.228
|
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chrX_-_132947141
|
1.222
|
|
GPC3
|
glypican 3
|
|
chr1_+_205293217
|
1.221
|
NM_001018053
NM_006212
|
PFKFB2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2
|
|
chr1_-_11909042
|
1.212
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr2_-_61618165
|
1.212
|
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr2_+_219751230
|
1.211
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr2_+_219751207
|
1.206
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr11_+_118463906
|
1.199
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chr7_-_27149750
|
1.197
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr2_+_17585287
|
1.197
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr5_+_70918856
|
1.194
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr9_+_70926205
|
1.189
|
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr7_-_23476497
|
1.181
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr2_+_17585462
|
1.180
|
|
VSNL1
|
visinin-like 1
|
|
chr6_-_138862271
|
1.172
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr6_+_80397690
|
1.168
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chr1_-_159257595
|
1.166
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr2_+_27518736
|
1.161
|
NM_001168364
NM_173853
|
KRTCAP3
|
keratinocyte associated protein 3
|
|
chr7_+_16652282
|
1.147
|
NM_001159767
NM_014038
|
BZW2
|
basic leucine zipper and W2 domains 2
|
|
chr17_-_21097170
|
1.144
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr8_+_55210333
|
1.140
|
NM_014175
|
MRPL15
|
mitochondrial ribosomal protein L15
|
|
chr1_+_61320213
|
1.138
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr12_-_63439449
|
1.137
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr17_-_7078304
|
1.136
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr16_-_15644465
|
1.128
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr3_-_50304706
|
1.127
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr19_-_53914768
|
1.125
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr19_+_40431618
|
1.123
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr20_+_43028533
|
1.122
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr6_+_41622111
|
1.119
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chrX_+_49531209
|
1.115
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr6_-_35996915
|
1.095
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr14_+_102458745
|
1.089
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr8_+_95722516
|
1.084
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr8_-_143482433
|
1.081
|
NM_145003
|
TSNARE1
|
t-SNARE domain containing 1
|
|
chr6_-_53517466
|
1.079
|
|
GCLC
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr17_-_7078446
|
1.074
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr9_-_106730256
|
1.073
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr6_+_32057779
|
1.071
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr19_-_44158130
|
1.069
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr10_+_101532450
|
1.068
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr19_+_40224457
|
1.067
|
|
HPN
|
hepsin
|
|
chr1_-_28432060
|
1.067
|
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr5_+_70918888
|
1.065
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr5_+_132237253
|
1.064
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr10_+_101532544
|
1.062
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr1_-_39097904
|
1.062
|
NM_022157
|
RRAGC
|
Ras-related GTP binding C
|
|
chr1_-_28432095
|
1.060
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr17_-_7078399
|
1.058
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr2_+_46779827
|
1.057
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr6_-_8047731
|
1.056
|
NM_001135650
NM_004280
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1
|
|
chr12_+_51686512
|
1.055
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr1_-_159459814
|
1.052
|
|
APOA2
|
apolipoprotein A-II
|
|
chr11_-_72141064
|
1.050
|
NM_001040118
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr2_+_219751192
|
1.050
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr2_+_241903395
|
1.045
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr14_+_19993204
|
1.038
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr12_-_94860504
|
1.036
|
NM_182496
|
CCDC38
|
coiled-coil domain containing 38
|
|
chr12_+_53005192
|
1.035
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr1_-_77921681
|
1.035
|
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr10_+_5478521
|
1.033
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr12_-_63439456
|
1.033
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_+_53005183
|
1.033
|
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr17_-_73636386
|
1.032
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr5_-_141372721
|
1.031
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr9_-_106730157
|
1.031
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr12_+_51686504
|
1.031
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr6_-_138862273
|
1.028
|
|
|
|
|
chr1_+_181422016
|
1.027
|
|
LAMC2
|
laminin, gamma 2
|
|
chr1_-_199635057
|
1.025
|
|
LAD1
|
ladinin 1
|
|
chr11_+_18300689
|
1.015
|
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa
|
|
chr19_+_38979502
|
1.014
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr10_-_32257768
|
1.014
|
NM_018287
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chrX_+_30581523
|
1.012
|
|
GK
|
glycerol kinase
|
|
chr8_+_95722575
|
1.010
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr2_+_206732575
|
1.006
|
|
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2
|
|
chr17_-_6495566
|
0.998
|
NM_016060
|
MED31
|
mediator complex subunit 31
|
|
chr8_+_11599119
|
0.998
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr1_-_152459697
|
0.997
|
NM_001098616
NM_015449
NM_138740
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr5_+_70918893
|
0.994
|
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr6_-_35996794
|
0.993
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr2_+_113119985
|
0.990
|
NM_005415
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1
|
|
chr1_+_9522107
|
0.981
|
NM_032315
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr12_-_63439415
|
0.981
|
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr1_-_199635240
|
0.980
|
NM_005558
|
LAD1
|
ladinin 1
|
|
chr6_+_7486862
|
0.980
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr12_+_53005177
|
0.978
|
NM_016057
|
COPZ1
|
coatomer protein complex, subunit zeta 1
|
|
chr6_+_7486836
|
0.975
|
|
DSP
|
desmoplakin
|
|
chr2_+_74610039
|
0.973
|
NM_013247
NM_145074
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr17_+_36636404
|
0.972
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr3_-_57088332
|
0.970
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr17_-_32730804
|
0.967
|
NM_198837
NM_198838
|
ACACA
|
acetyl-CoA carboxylase alpha
|
|
chr1_-_152797707
|
0.966
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr1_-_152797760
|
0.966
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|