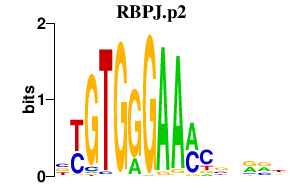
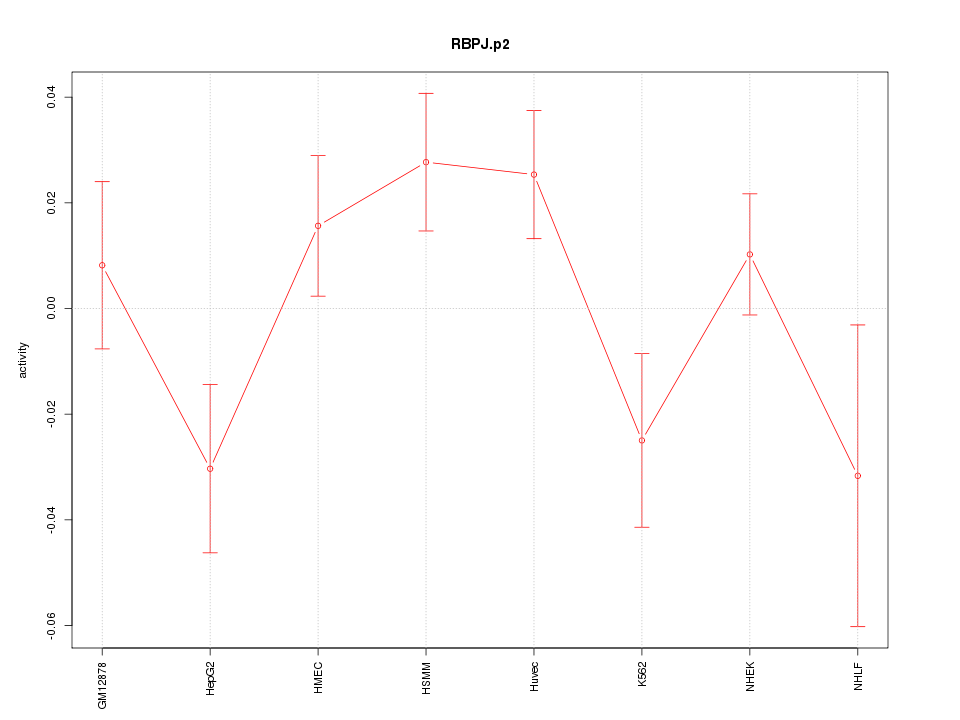
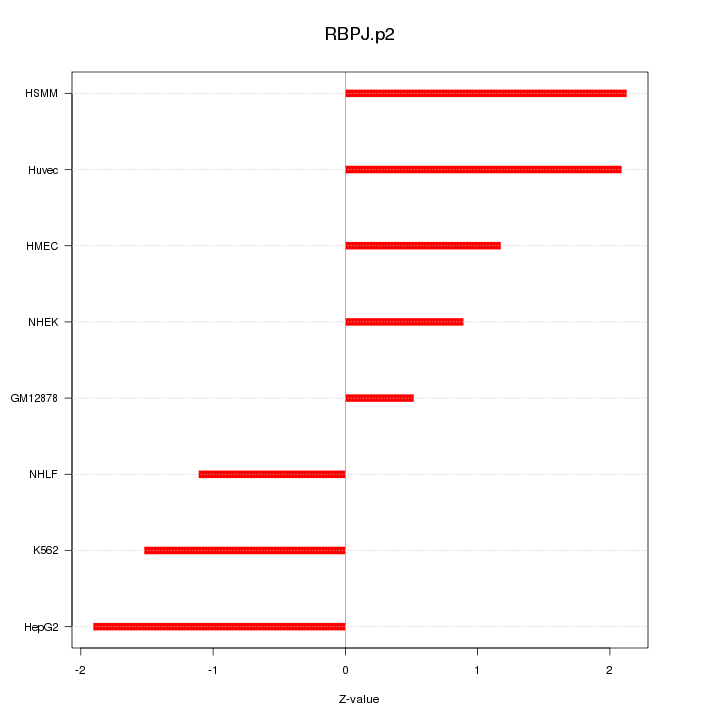
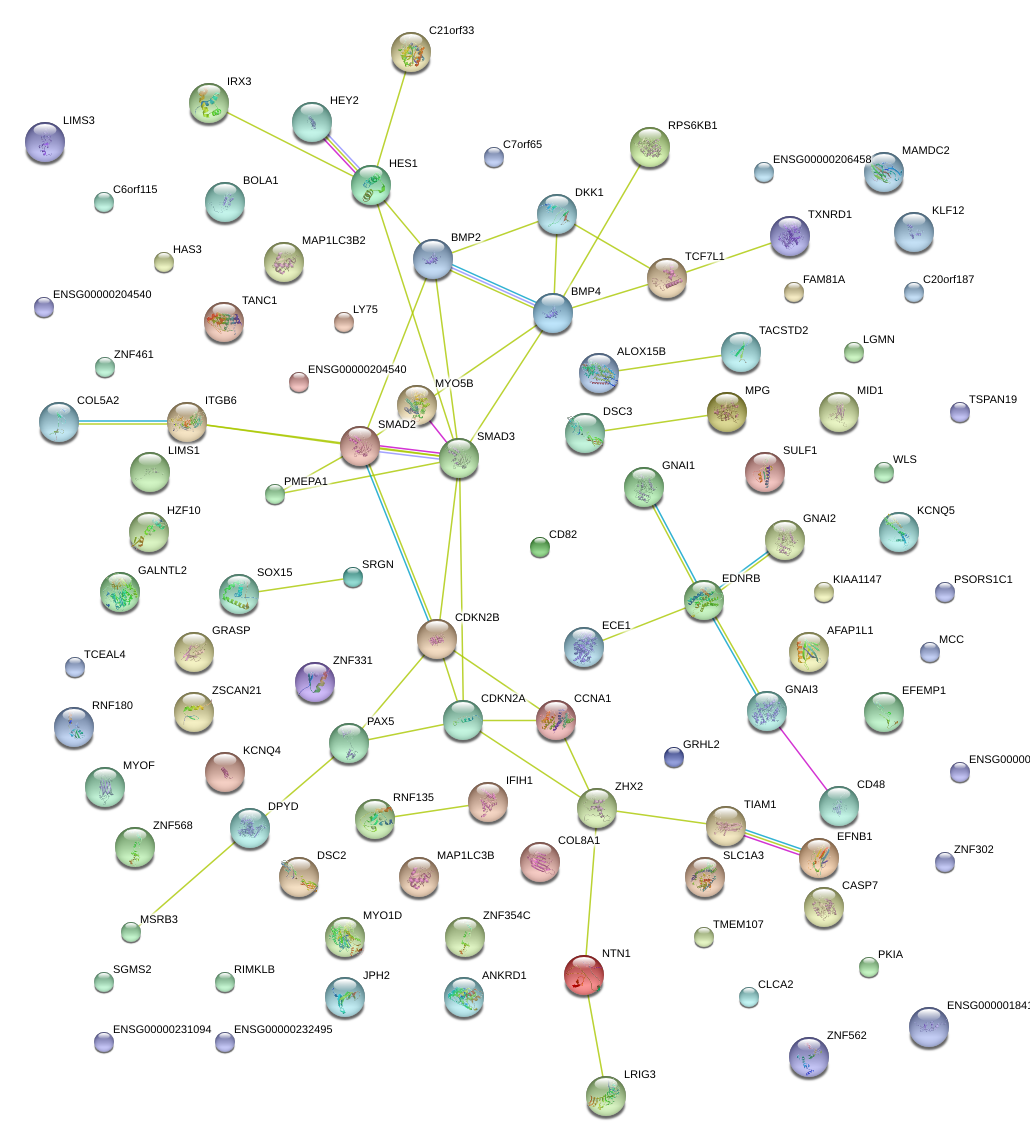
|
chr19_-_39859997
|
2.058
|
|
|
|
|
chr19_+_39860398
|
2.009
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr10_-_95231933
|
1.736
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr22_+_25398690
|
1.606
|
|
LOC388889
|
hypothetical LOC388889
|
|
chr3_+_100840129
|
1.462
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr1_-_68470585
|
1.458
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr13_-_73606362
|
1.384
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr2_+_159533329
|
1.330
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr9_+_71848555
|
1.329
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr8_+_79590890
|
1.224
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr8_+_79591074
|
1.169
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chrX_+_67965517
|
1.168
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr21_-_31853160
|
1.163
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr14_-_92284626
|
1.150
|
NM_001008530
NM_005606
|
LGMN
|
legumain
|
|
chr2_-_189752575
|
1.145
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr17_+_7883076
|
1.133
|
NM_001039130
NM_001039131
NM_001141
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B
|
|
chr20_+_6696744
|
1.118
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chr15_+_57517564
|
1.000
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr1_-_98159113
|
0.950
|
|
DPYD
|
dihydropyrimidine dehydrogenase
|
|
chr20_-_55718351
|
0.945
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr3_+_195336631
|
0.944
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr8_+_70567634
|
0.941
|
|
SULF1
|
sulfatase 1
|
|
chr2_-_160764819
|
0.938
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr10_-_92670764
|
0.924
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr15_+_65205107
|
0.854
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr6_+_31190505
|
0.839
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr10_+_115429622
|
0.827
|
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr12_-_57600336
|
0.819
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr12_+_8741746
|
0.817
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr14_-_53491019
|
0.803
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr6_+_73388291
|
0.801
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr8_+_70567581
|
0.786
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr18_-_26876622
|
0.770
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr9_-_21984379
|
0.742
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr12_-_57600564
|
0.735
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr5_-_112658456
|
0.729
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr1_+_148137778
|
0.699
|
NM_016074
|
BOLA1
|
bolA homolog 1 (E. coli)
|
|
chr17_+_26322081
|
0.692
|
NM_001184992
NM_032322
NM_197939
|
RNF135
|
ring finger protein 135
|
|
chr1_+_86662356
|
0.681
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr6_+_139055244
|
0.679
|
|
FLJ46906
|
hypothetical LOC441172
|
|
chr10_+_115429415
|
0.678
|
NM_001227
NM_033338
NM_033339
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr20_+_55718718
|
0.667
|
|
|
|
|
chr2_-_160469438
|
0.665
|
|
LY75-CD302
LY75
|
LY75-CD302 read-through transcript
lymphocyte antigen 75
|
|
chr9_-_21984329
|
0.655
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr2_-_56004366
|
0.649
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr4_+_109033868
|
0.644
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_178419938
|
0.640
|
NM_014594
|
ZNF354C
|
zinc finger protein 354C
|
|
chr16_-_52877868
|
0.636
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr17_-_28228184
|
0.626
|
|
MYO1D
|
myosin ID
|
|
chr9_-_21984414
|
0.617
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr20_+_55718619
|
0.613
|
|
|
|
|
chr2_-_162883062
|
0.611
|
NM_022168
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr7_+_47661366
|
0.610
|
NM_001123065
|
C7orf65
|
chromosome 7 open reading frame 65
|
|
chr13_+_31318828
|
0.607
|
|
EEF1DP3
|
eukaryotic translation elongation factor 1 delta pseudogene 3
|
|
chr7_+_79602023
|
0.606
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr10_+_70517863
|
0.604
|
|
SRGN
|
serglycin
|
|
chr12_+_115481568
|
0.599
|
NM_001085481
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr3_+_64645819
|
0.596
|
|
LOC100507098
|
hypothetical LOC100507098
|
|
chr2_-_160469512
|
0.592
|
NM_001198759
NM_001198760
NM_002349
|
LY75-CD302
LY75
|
LY75-CD302 read-through transcript
lymphocyte antigen 75
|
|
chr13_-_77391525
|
0.592
|
|
|
|
|
chr17_-_7434113
|
0.591
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr2_+_108637916
|
0.585
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr17_+_26322168
|
0.582
|
|
RNF135
|
ring finger protein 135
|
|
chr1_-_58815753
|
0.577
|
NM_002353
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr16_-_52877829
|
0.576
|
|
IRX3
|
iroquois homeobox 3
|
|
chr8_+_102573837
|
0.572
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr8_+_102574012
|
0.571
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr5_+_148631636
|
0.566
|
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr5_+_63497408
|
0.558
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr12_+_103133631
|
0.557
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr2_+_108637698
|
0.554
|
NM_001193485
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chrX_+_102727068
|
0.554
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr5_+_148631588
|
0.552
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr16_-_52877467
|
0.551
|
|
IRX3
|
iroquois homeobox 3
|
|
chr2_-_160765008
|
0.541
|
|
ITGB6
|
integrin, beta 6
|
|
chr7_-_141048358
|
0.541
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chr19_+_58715988
|
0.540
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr19_-_41849558
|
0.535
|
NM_153257
|
ZNF461
|
zinc finger protein 461
|
|
chr13_-_77390907
|
0.532
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chr16_+_67698943
|
0.531
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr13_+_35904408
|
0.530
|
NM_001111045
NM_003914
|
CCNA1
|
cyclin A1
|
|
chr11_+_44543716
|
0.529
|
NM_001024844
NM_002231
|
CD82
|
CD82 molecule
|
|
chr10_+_53744043
|
0.522
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr17_+_8865547
|
0.516
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr12_+_63958949
|
0.513
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr1_-_21478571
|
0.513
|
NM_001113347
|
ECE1
|
endothelin converting enzyme 1
|
|
chr2_+_85213884
|
0.511
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr2_+_110013297
|
0.510
|
NM_033514
|
LIMS3-LOC440895
LIMS3
|
LIMS3-LOC440895 read-through
LIM and senescent cell antigen-like domains 3
|
|
chr12_-_83954185
|
0.504
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr5_+_36644187
|
0.496
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_+_44543788
|
0.494
|
|
CD82
|
CD82 molecule
|
|
chr6_+_126112506
|
0.490
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr18_-_45630154
|
0.477
|
|
MYO5B
|
myosin VB
|
|
chr1_-_158948176
|
0.473
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr3_+_16191159
|
0.466
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr19_+_42099073
|
0.465
|
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr12_+_50686990
|
0.460
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr1_-_58815415
|
0.456
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chrX_-_10548458
|
0.455
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr8_+_123863169
|
0.455
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr20_-_42249146
|
0.451
|
|
JPH2
|
junctophilin 2
|
|
chr6_+_139391511
|
0.446
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr19_+_42033106
|
0.441
|
NM_003419
|
ZNF345
|
zinc finger protein 345
|
|
chr5_-_94443300
|
0.433
|
NM_001002796
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr12_+_83954229
|
0.428
|
NM_001079910
NM_032165
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr3_-_150858274
|
0.428
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr8_-_98359366
|
0.423
|
|
TSPYL5
|
TSPY-like 5
|
|
chr6_+_49626431
|
0.423
|
|
|
|
|
chr8_-_28403652
|
0.420
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr19_-_42098758
|
0.416
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr1_+_216585298
|
0.415
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr18_+_18969724
|
0.412
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr11_-_6298284
|
0.410
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr12_-_102413875
|
0.409
|
NM_198521
NM_001099336
|
C12orf42
|
chromosome 12 open reading frame 42
|
|
chr7_-_44992729
|
0.408
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr8_-_98359248
|
0.405
|
|
TSPYL5
|
TSPY-like 5
|
|
chr21_-_14677310
|
0.403
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr17_+_71232370
|
0.402
|
NM_001005619
|
ITGB4
|
integrin, beta 4
|
|
chr16_+_56219802
|
0.398
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr4_-_55686503
|
0.393
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chrX_+_106956158
|
0.389
|
|
MID2
|
midline 2
|
|
chr17_+_18700385
|
0.389
|
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chr3_+_42107711
|
0.386
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr19_+_42689881
|
0.384
|
|
ZNF793
|
zinc finger protein 793
|
|
chr11_+_66268817
|
0.379
|
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr6_+_126112431
|
0.376
|
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr11_+_66268740
|
0.373
|
NM_024650
|
C11orf80
|
chromosome 11 open reading frame 80
|
|
chr3_+_103029523
|
0.370
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr3_-_145049853
|
0.370
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr10_+_75343352
|
0.370
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr4_-_83938920
|
0.368
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr10_-_33663984
|
0.368
|
|
NRP1
|
neuropilin 1
|
|
chr8_-_98359347
|
0.367
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr9_-_5329745
|
0.366
|
NM_006911
|
RLN1
|
relaxin 1
|
|
chr2_-_31293882
|
0.366
|
NM_001145122
|
CAPN14
|
calpain 14
|
|
chr5_+_42792676
|
0.363
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr12_-_48387233
|
0.361
|
|
|
|
|
chr4_+_146623405
|
0.361
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr12_-_93921586
|
0.361
|
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr6_+_126112417
|
0.360
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr8_+_123863224
|
0.360
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr11_-_5662762
|
0.358
|
NM_033034
NM_033092
NM_033093
|
TRIM5
|
tripartite motif containing 5
|
|
chr2_+_69093641
|
0.356
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr6_+_49626317
|
0.355
|
|
C6orf141
|
chromosome 6 open reading frame 141
|
|
chr2_-_175255717
|
0.354
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr12_-_69317441
|
0.354
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr2_+_237143118
|
0.352
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr9_+_17124974
|
0.352
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr22_+_28493351
|
0.349
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr3_+_160964574
|
0.349
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr19_+_61681294
|
0.347
|
|
LOC100128252
|
hypothetical LOC100128252
|
|
chr9_+_22436783
|
0.346
|
NM_022160
|
DMRTA1
|
DMRT-like family A1
|
|
chr19_-_42098919
|
0.346
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr1_-_226202210
|
0.345
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr12_-_93921618
|
0.345
|
NM_018838
|
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
|
|
chr4_+_146622154
|
0.344
|
NM_005900
|
SMAD1
|
SMAD family member 1
|
|
chr9_+_6403321
|
0.341
|
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr19_-_55220388
|
0.340
|
|
VRK3
|
vaccinia related kinase 3
|
|
chr11_-_58102169
|
0.339
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr11_-_60885928
|
0.338
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr22_-_45311730
|
0.337
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr3_+_49002774
|
0.336
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr8_-_102031943
|
0.333
|
NM_001135702
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr5_-_156935283
|
0.333
|
|
|
|
|
chr19_-_58449752
|
0.328
|
|
ZNF677
|
zinc finger protein 677
|
|
chr12_+_94391952
|
0.326
|
NM_006838
|
METAP2
|
methionyl aminopeptidase 2
|
|
chr11_-_60885849
|
0.322
|
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr10_+_70517823
|
0.319
|
NM_002727
|
SRGN
|
serglycin
|
|
chr15_-_65334022
|
0.318
|
NM_024666
|
AAGAB
|
alpha- and gamma-adaptin binding protein
|
|
chr2_+_87536174
|
0.318
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr19_+_42689680
|
0.317
|
NM_001013659
|
ZNF793
|
zinc finger protein 793
|
|
chr14_-_59407103
|
0.317
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr13_-_40138607
|
0.317
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr1_+_199975563
|
0.315
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr6_+_139391570
|
0.315
|
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr14_+_51025604
|
0.314
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr2_-_191724169
|
0.312
|
NM_003151
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr15_+_76889873
|
0.311
|
|
MORF4L1
|
mortality factor 4 like 1
|
|
chr11_-_87548199
|
0.309
|
NM_022337
|
RAB38
|
RAB38, member RAS oncogene family
|
|
chr10_-_92671005
|
0.309
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr5_+_14493905
|
0.306
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr7_-_19715127
|
0.305
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr2_-_191723951
|
0.304
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr14_+_72773957
|
0.304
|
NM_173462
|
NDRG2
PAPLN
|
NDRG family member 2
papilin, proteoglycan-like sulfated glycoprotein
|
|
chr1_-_145699308
|
0.303
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr3_+_114414064
|
0.303
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr3_+_49002285
|
0.297
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr12_-_48387459
|
0.296
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr4_-_84475036
|
0.296
|
NM_001098540
|
HPSE
|
heparanase
|
|
chr17_+_35727998
|
0.295
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr14_+_72672960
|
0.295
|
|
PSEN1
|
presenilin 1
|
|
chr2_-_191723996
|
0.294
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr4_+_61749540
|
0.292
|
|
LPHN3
|
latrophilin 3
|
|
chr14_+_72672913
|
0.292
|
|
PSEN1
|
presenilin 1
|
|
chr5_+_94981735
|
0.291
|
NM_199243
|
GPR150
|
G protein-coupled receptor 150
|
|
chr1_-_23393808
|
0.290
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr4_-_84475328
|
0.290
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr21_-_17906975
|
0.288
|
|
BTG3
|
BTG family, member 3
|
|
chr2_+_87536106
|
0.288
|
|
NCRNA00152
|
non-protein coding RNA 152
|
|
chr19_-_55220439
|
0.287
|
|
VRK3
|
vaccinia related kinase 3
|